Figure 3. Transient maternal DMRs target a divergent set of CpG island promoters.
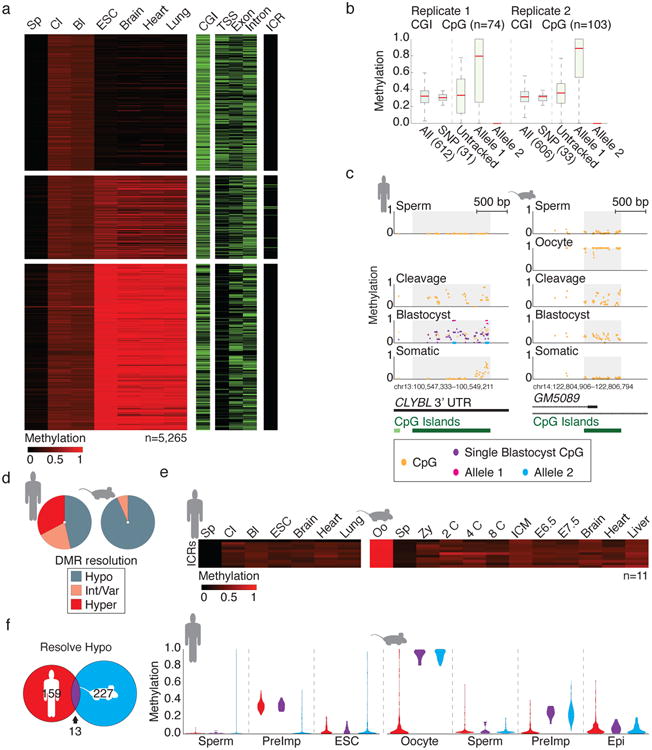
a. Heatmap of 5,265 100 bp tiles consistent with maternally contributed monoallelic methylation (Methods). Tiles are partitioned according to their hypo (≤0.2), intermediate/variable (0.2<x<0.8) or hypermethylated (≥0.8) resolution in ESCs. Feature annotations are included as separate heatmaps.
b. Boxplots of CGI DMR methylation for two independent single blastocysts, with heterozygous SNP-linked CpGs highlighted. Within each replicate, 31 and 33 CGI DMRs contain CpGs that could be assigned to parental loci. In each case, DNA methylation is restricted to only one of the two alleles. Untracked refers to the inferred methylation status prior to haplotype segregation. Red line signifies the median, boxes and whiskers the 25th/75th and 2.5th/97.5th percentiles.
c. Single CpG track of a conserved preimplantation-specific DMR in human and mouse. Human blastocyst data includes information from the pooled sample as well as for a single blastocyst replicate (purple) with allele-tracked methylation for 10 CpGs highlighted in pink and blue. Annotated CGIs are included below.
d. Resolution of CGIs that behave as maternal DMRs in human and mouse.
e. Heatmap of orthologous ICRs over human and mouse preimplantation development.
f. Orthologous hypomethylation-resolving CGID MRs in human and mouse share only 13 equivalently regulated regions. When methylation values of mouse or human specific DMRs are tracked in the alternate species, they are constitutively hypomethylated, indicating that oogenesis targets equivalent genomic features but at species-specific sequences. PreImp refers to the average value for cleavage and blastocyst in human or 8 cell and ICM in mouse.
