Extended Data Figure 6. Genomic characterization of transient maternally contributed imprint-like regions.
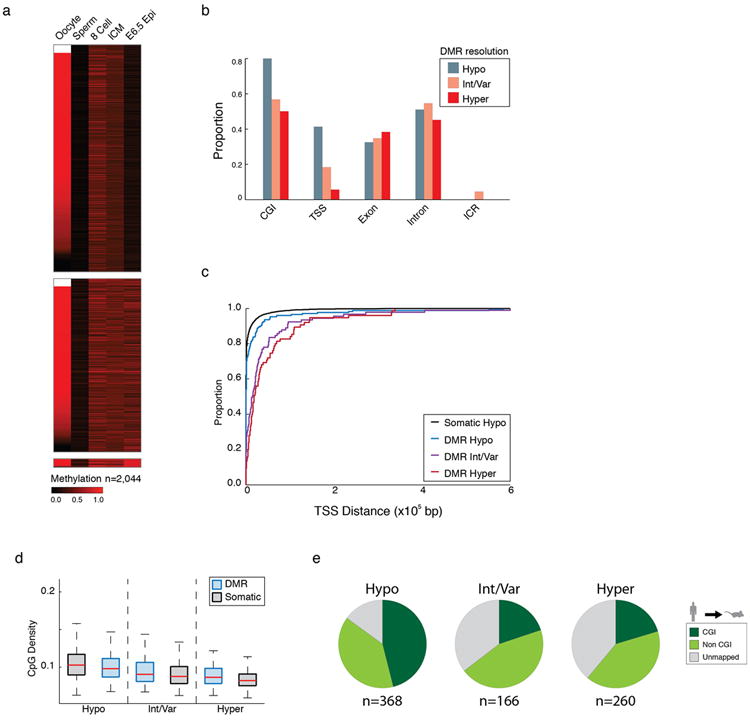
a. Heatmap of 100 bp tiles in mouse preimplantation identified using the same criteria as applied to human (Methods). This criteria, which assumes limited de novo methylation, identifies 2,044 tiles in mouse where methylation is ≥ 0.2 in both 8 cell and the ICM, there is ≥0.2 methylation difference between the ICM and sperm, and this difference is significant via t test, (q-value < 0.05). 89% of those tiles that are captured in the mouse oocyte are monoallelically inherited and show significant differences between the gametes by t-test, providing an empirical upper bound on the False Discovery Rate for this strategy when applied to human of ≤ 0.11, assuming the underlying principles of imprint regulation are the same as in mouse.
b. The proportion of 100 bp tiles, classified according to their resolution in ESCs, for each genomic feature presented in Figure 3a.
c. Cumulative density function (CDF) plot of the distance to the nearest annotated TSS for CGI DMRs that resolve to hypomethylation, intermediate/variable methylation, or hypermethylation. There is a discrepancy in genomic location between those that resolve to hypomethylation, of which a sizable fraction are in the TSS, and those that do not, which are generally enriched further downstream.
d. Boxplots of CpG density for CGI DMRs that resolve to hypomethylation, intermediate/variable methylation, or hypermethylation paired with comparable non-DMR CGIs (Somatic). Those resolving to hypomethylation have higher CpG densities than those that resolve to intermediate/variable or hypermethylation, but have slightly lower CpG density than non-DMR, constitutively hypomethylated CGIs. Alternatively, while CGIs that resolve to hypermethylation show a lower CpG density than other DMRs, they show higher density than non-DMR hypermethylated islands, suggesting some level of protection against deamination as an attribute of their uniquely hypomethylated status in the male germline.
e. Pie charts of cross species alignment and CGI status of human CGI DMRs into mouse. Those that resolve to hypomethylation are more often conserved in mouse and more frequently retain their CGI status, whereas those resolving to hypermethylation are less conserved. Moreover, intermediate/variable and hypermethylation-resolving regions that do align are less frequently retained as CGIs suggesting that hypomethylation specific to the male germline is insufficient to protect these regions from progressive deamination over time. 368, 166, and 260 CGIs comprise the hypo, intermediate/variable, and hyper methylation sets, respectively,
