Abstract
Sulfated beta-O4 lignin (SbO4L), a non-saccharide glycosaminoglycan mimetic, was recently disclosed as a novel exosite 2-directed thrombin inhibitor with capability of mimicking sulfated tyrosine sequences of glycoprotein Ibα resulting in dual anticoagulant and antiplatelet activities. SbO4L engages essentially the same residues of exosite 2 as heparin and yet induces allosteric inhibition. Fluorescence spectroscopic studies indicate that SbO4L reduces access of the active site to molecular probes and affinity studies at varying salt concentrations nearly 6 ionic interactions, similar to heparin, but much higher non-ionic contribution. The results suggest that subtle increase in non-electrostatic forces arising from SbO4L’s aromatic scaffold appear to be critical for inducing allosteric dysfunction of thrombin’s active site.
Keywords: allosteric inhibition, anticoagulants, coagulation, glycosaminoglycan mimetics, thrombin
Introduction
Hemostasis is a fine-tuned process that maintains a delicate balance between bleeding and clotting through a dynamic sequence of events involving soluble molecules, cells and sub-vasculature factors. A key aspect of this process is the blood coagulation cascade, which relies on highly selective recognition of macromolecular substrates by their proteases [1–3]. Selectivity for most enzyme – substrate pairs arises from complementarity of three-dimensional features around the active site, but that for coagulation proteases appears to rely on allosteric sites [2–4]. For example, thrombin can function either as a procoagulant or an anticoagulant depending on whether its macromolecular partner is fibrinogen or thrombomodulin [4,5].
Thrombin is arguably the most important coagulation protease interacting with many proteins including pro-cofactors and cell surface receptors to effect its function. In addition to its one-of-a-kind active site, it is endowed several exosites including the sodium binding site, and anion-binding exosites 1 and 2 [6–7]. The three exosites are located several angstroms away from the active site and modulate thrombin’s catalytic activity. Recent studies indicate that thrombin is a mobile protease exhibiting multiple conformations that can be differentially stabilized by using appropriate exosite binding ligands [6,8]. Molecular forces that contribute to these processes are just beginning to be unraveled.
Understanding thrombin allosterism is important because of the possibility of developing anticoagulants for treating thrombotic disorders and procoagulants for treating hemophilic disorders. A priori, allosterism offers regulatory advantages that orthosterism cannot [4,9,10]. For example, allosteric inhibitors can be designed to maximally inhibit less than 100%, as shown earlier [11], which may be of special value when complete inhibition results in deleterious consequences. Considering that thrombin is a highly plastic enzyme, understanding forces contributing to allosteric networks may help develop novel inhibitors with potentially reduced bleeding complications.
Recently, we have developed sulfated β-O4 lignin (SbO4L), a sulfated non-saccharide glycosaminoglycan mimetic (NSGM), as a macromolecular allosteric inhibitor of thrombin [12]. SbO4L induced a reduction in the catalytic efficiency of thrombin by binding to several positively charged residues of exosite 2. This was in striking contrast to heparin, which also binds exosite 2 with similar set of residues, and yet does not induce catalytic dysfunction [13]. Further, the allosteric disruption of catalysis by SbO4L occurred only for thrombin suggesting a highly selective process. In contrast, heparin binds to several coagulation proteases and demonstrates essentially no selectivity [14]. Finally, SbO4L was found to compete with glycoprotein Ibα (GPIbα) of platelets in binding to exosite 2 of thrombin and thereby exert dual anticoagulant and antiplatelet activities. SbO4L is the first synthetic macromolecule to exhibit such a mechanism and offers an interesting platform to study the nature of allosterism and its dependence on the structure of effector ligand. This work discusses studies focused on understanding why SbO4L induces catalytic dysfunction and deciphering similarities and differences between thrombin interactions with SbO4L and heparin. The results provide new avenues to explore in developing sulfated NSGMs as allosteric regulators of thrombin.
Materials and Methods
Materials
Active site labeled fluorescein-FPR-thrombin (fFPR-thrombin) was purchased from Haematologic Technologies (Essex Junction, VT). Fluorescence measurements were recorded using a QM4 spectrofluorometer (Photon Technology International, Birmingham, NJ). All other reagents were reagent grade and purchased from either Sigma Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA).
Fluorescence Quenching Studies
Quenching studies using a collisional quencher were performed in 20 mM Tris-HCl buffer containing 100mM NaCl, 2.5 mM CaCl2, and 0.1% PEG 8000 at pH 7.4. A mixture of fFPR-thrombin (170 nM) and 44 μg/mL SbO4L (or buffer) was titrated with increasing concentrations of acrylamide (10 M stock, 1 μL additions) and fluorescence intensity at 522 nm (λEX = 490 nm) was monitored at 25°C. Excitation and emission slit widths were 1 mm each. The ratio of the fluorescence without the quencher (F0) to that in the presence of the quencher (F) was plotted against the quencher concentration (Q), which yielded a linear relationship that could be fitted using the Stern-Volmer equation 1 [15]. In this equation, Q is the concentration of the quencher, KSV is the Stern-Volmer constant and is equal to kqτ0 (kq = bimolecular quenching constant, τ0 = fluorophore lifetime in the absence of the quencher).
| (Eq. 1) |
Binding Affinity Studies
The affinity of SbO4L for fFPR-thrombin was measured in 20 mM Tris-HCl, pH 7.4, containing varying concentrations of NaCl (100–200 mM), 2.5 mM CaCl2 and 0.1% PEG 8000. A concentration of 170 nM fFPR-thrombin was sufficient to give good fluorescence emission signal at 522 nm (λEX = 490 nm) at 25°C. Excitation and emission slit widths were 1 mm each. Titrations were performed by adding small aliquots of concentrated stock of SbO4L so as to not increase the total volume of the sample by more than 5% and monitoring the change in fluorescence at 522 nm. This change in fluorescence signal was fitted using the classic quadratic binding equation 2 to calculate the dissociation constant KD at various concentrations of salt. IN this equation, ΔF represents the change in fluorescence of fFPR-thrombin at each addition of SbO4L from the initial fluorescence F0 and ΔFMAX represents the maximal change in fluorescence observed when the enzyme is saturated with the inhibitor. The binding stoichiometry was assumed to be 1:1 for SbO4L – thrombin complex.
| (Eq. 2) |
To resolve contribution of the ionic and non-ionic forces to the SbO4L–thrombin interaction, equation 3 was used. The slope (Z×Ψ) of the line correlates the sensitivity of the binding affinity to salt concentration, while the intercept corresponds to binding affinity when all ionic interactions are completely screened ([Na+] = 1 M).
| (Eq. 3) |
Results
Exosite 2 Binding by SbO4L Reduces Steric Access to Thrombin’s Active Site
To assess the steric accessibility of thrombin’s active site upon SbO4L binding, quenching studies were carried out on fFPR-thrombin using a collisional quencher. We reasoned that if SbO4L binding to exosite 2 induces significant changes in the conformational state of the active site, then a collisional quencher would affect fluorescence of the active site probe at a rate different from that for the native enzyme. Acrylamide and iodide are two collisional quenchers typically used in the literature for this purpose with good success [15]. Both molecules quench protein’s fluorescence through direct molecular contact, which implies that the method senses steric and electrostatic environment in the vicinity of the fluorophore. Figure 1A shows the acrylamide-induced quenching of fluorescein fluorescence of fFPR-thrombin in free and SbO4L bound forms. At low levels of acrylamide (0.04 – 0.12 M), a rapid decrease in fluorescence was observed, which gradually reached a plateau at acrylamide concentrations of 0.56 M, for enzyme alone. However, in the presence of saturating concentrations of SbO4L, fluorophore quenching was significantly retarded such that at 0.56 M acrylamide, the fluorescence of the complex was significantly higher than the thrombin alone. Further, a bolus of SbO4L (44 μM final concentration) added thrombin pre-quenched with 0.56 M acrylamide resulted in a nearly complete recovery of fluorescence to levels observed in the titration of SbO4L – thrombin complex (not shown).
Figure 1.
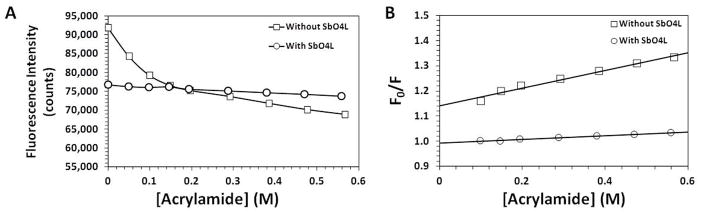
(A) Fluorescence of fFPR-thrombin with and without SbO4L at various concentrations of collisional quencher acrylamide. (B) Stern-Volmer analysis of the fluorescence quenching data. Solid line represents linear regression analysis using equation 1.
Collisional quenching can be quantitatively analyzed in terms of one or more species of fluorophores present on the protein using various forms of Stern-Volmer relationships [15]. Figure 1B shows Stern-Volmer plots that can be explained well using a linear relationship (Eq. 1) suggesting the presence of only one species of fluorophore for both free thrombin and thrombin – SbO4L complex. The slope of the linear regression corresponds to the quenching constant (KSV), which was calculated to be 0.351 for free thrombin and 0.073 for the complex, a difference of almost 4.8-fold. This suggests that the allosteric conformational change induced by SbO4L in thrombin reduces the significant accessibility of the fluorophore to acrylamide. Considering that the fluorescein fluorophore is located almost at the P4 site (P4fFPRP1), these results indicate that the allosteric change induced by SbO4L is extensive and is not just limited to the catalytic triad.
SbO4L – Thrombin Interaction is Primarily Electrostatic, but with Significant Non-Ionic Component
To determine the nature of interactions made by SbO4L with thrombin, the dissociation constant of complex (KD,OBS) was measured as a function of NaCl concentration. Fluorescence titrations were performed by monitoring the change in fluorescence of fFPR-thrombin with increasing concentrations of SbO4L (Figure 2A), which could be fitted by quadratic binding equation 2 to calculate the affinity of the complex. A significant loss in binding affinity was observed for a relatively small change in the concentration of NaCl (100 to 200 mM) suggesting that the interaction was highly sensitive to ionic strength of the buffer. To resolve the forces contributing to the interaction, a plot of logKD,OBS versus log[Na+] was prepared following literature reports on the application of the protein-polyelectrolyte interaction theory [16,17]. According to this theory, binding of a polyelectrolyte, such as SbO4L, to a protein, such as thrombin, in the presence of a monovalent salt can be considered to be an ion exchange-type process involving release of bound counter-ions from SbO4L. The theory predicts that the overall binding energy, as obtained from KD,OBS, will be comprised of ionic (KD,I) and non-ionic (KD,NI) components, which can be obtained from the intercept and slope of the logKD,OBS versus log[Na+] plot, respectively.
Figure 2.
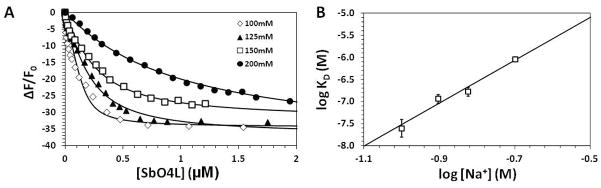
(A) Fluorescence titration of fFPR-thrombin as a function of SbO4L in pH 7.4 buffer containing varying levels sodium chloride. Solid line shows non-linear regression analysis using equation 2 to obtain the KD,OBS of the interaction. (B) A profile of logKD versus log[Na+] for SbO4L – thrombin interaction to parse ionic and non-ionic components of binding energy using equation 3.
Application of the protein – polyelectrolyte theory using equation 3 gave a slope of 4.85 ± 0.43 and an intercept of −2.67 ± 0.33 for SbO4L – thrombin system (Figure 2B). Assuming that formation of each ionic bond for such system results in the release of 0.8 Na+ ions, as reported in the literature [16,18], the slope corresponds to formation of approximately 6.1 ± 0.5 salt bridges between SbO4L and thrombin. This is approximately equal to the ionic interactions noted for other exosite 2 binding ligands such as heparin, thrombomodulin and GPIbα (Table 1) [16,19–21]. This implies that a trimeric SbO4L unit with 2 sulfate groups per monomer would be the simplest scaffold capable of recognizing thrombin with high selectivity. The structure of SbO4L shows that majority of β-O4 linked monomers can possess 2 sulfate groups. These results should assist with designing advanced homogenous molecules based on the SbO4L scaffold.
Table 1.
Comparison of forces involved in the interactions of thrombin with allosteric ligands.
| Ligand | Z × Ψ | ΔGionica | KD,nonionic | ΔGnonionicb | Reference |
|---|---|---|---|---|---|
| kcal/mol | mM | kcal/mol | |||
| Hirudin peptide (Hir55–65) | 1.06 | −1.19c | nad | na | 19 |
| Thrombomodulin with CSe | 4.8 | −5.77 | 0.0125 | −6.66 | 19 |
| Thrombomodulin without CS | 2.2 | −2.34 | 0.000708 | −8.36 | 19 |
| Glycocalicin (Soluble GPIbα) | 4.2 | −4.73 | 2.88 | −3.45 | 20 |
| Glycocalicin (Soluble GPIbα) | 4.6 | −5.17c | na | na | 21 |
| Heparin | 4.8 | −5.4 | 53.3 | −1.73 | 16 |
| SbO4L | −4.85 ±0.43 f | −5.45 | 2.14 ±0.26 | −3.64 | this work |
free energy of binding at 150 mM NaCl arising from ionic interactions.
free energy of binding at 1 M NaCl arising from non-ionic interactions.
calculated from slope measured by authors.
not available.
chondroitin sulfate.
standard error obtained from linear regression.
The intercept of the double log plot corresponds to logKD,NI, which was calculated to be 2.14 ± 0.26 mM. Thus, the non-ionic binding energy component (ΔGNI) in SbO4L – thrombin interaction is approximately −3.64 kcal/mol, which represents 40.1% of the total binding energy under physiologically relevant salt concentrations (ΔGOBS = −9.09 kcal/mol). The remaining energy arises from ionic interactions and corresponds to approximately 60% contribution. Thus, the interaction between SbO4L and thrombin is primarily electrostatic. In comparison, heparin displays a higher electrostatic contribution of ~80% [16], while that for GPIbα is closer to SbO4L at ~58% [20]. Likewise, studies with thrombomodulin containing chondroitin sulfate shows ~47% electrostatic contribution, which reduces to approximately 22% for thrombomodulin without chondroitin sulfate [19].
Discussion
The plasticity of thrombin is regulated by a number of allosteric binding ligands [5–8]. The GPIbα – thrombin interaction has gained considerable interest over the years because of the the ambiguity surrounding whether exosite 1 or 2 or both form key contact point(s) [22,23]. But another interesting aspect of this interaction is that GPIbα allosterically induces reduction in the catalytic efficiency of thrombin, while also being essential for platelet activation [20,21,24]. Individually, the two processes are thus counter-productive. Whereas thrombin inhibition is anticoagulant, platelet activation is procoagulant. We had reasoned that these two seemingly opposing effects could be streamlined to a uniform anticoagulant effect by developing agents that compete with GPIbα in binding to thrombin while simultaneously inducing allosteric inhibition. This led to the design of SbO4L, which was chemically synthesized in only three high yielding steps, and shown to cause binding to a select group of exosite 2 residues and inhibit thrombin [12]. Despite the considerable background, questions remained on the nature of binding and allostery. These were also especially important to understand because exosite 2 ligands, which are mostly glycosaminoglycans including heparin and chondroitin sulfate, do not induce inhibition [13].
This work shows that the binding of SbO4L to thrombin significantly reduces the access of the active site to small fluoroprobes, which suggests that the entry of a thrombin substrate would be prevented. The extent of conformational change brought about by SbO4L appears to be fairly large because the fluorescein group is expected to be localized near the S4 subsite, which is several angstroms away from the Ser195 of the catalytic triad. Yet, these results do not convey exact information on the state of Ser195 in the bound complex. It is likely that the extensive conformational change around the active site extrapolates to Ser195 also but this needs to be shown more rigorously.
A second important point brought out in this study is that SbO4L – thrombin interaction more closely mimics GPIbα – thrombin interaction than heparin – thrombin interaction. Although all three molecules appear to be utilizing the same number of ionic interactions (i.e., 5 – 6), the contribution of non-ionic forces to interaction is more similar for GPIbα and SbO4L. The quantitative difference between heparin and GPIbα /SbO4L systems appears to be only about 20%, but the functional consequences are pretty dramatic. While SbO4L and GPIbα directly inhibit thrombin [12,20,24], heparin does not [13]. The atomistic reason for this difference is unclear at this time but it is instructive to note that both SbO4L and GPIbα rely on sulfated aromatic structures to recognize thrombin. In contrast, heparin is a polysaccharide and completely devoid of aromatic rings. Thus, it is likely that aromaticity in both these molecules plays a subtle role to enhance non-ionic interactions. Recent work with sulfated aromatic molecules has shown that exosite 2 contains multiple hydrophobic subsites [25,26] and one or more of these subsites may be important for inducing allosteric catalytic dysfunction.
In conclusion, we have identified that engaging exosite 2 of thrombin with a strongly negatively charged, but aromatic, molecule can induce direct and fairly extensive conformational change in the active site. This conformational change is the basis for enzyme inhibition.
Highlights.
Sulfated beta-O4 lignin (SbO4L) binding to exosite 2 restricts access of thrombin’s active site to molecular probes.
SbO4L forms 5 to 6 ionic interactions with exosite 2
Despite similarity with heparin, subtle differences in non-ionic forces induce thrombin inhibition by SbO4L.
Acknowledgments
This work was support by grants R01 HL90586 and P01 HL107152 from the National Heart, Lung and Blood Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Krishnaswamy S. The transition of prothrombin to thrombin. J Thromb Haemost. 2013;11(Suppl 1):265–276. doi: 10.1111/jth.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Page MJ, Macgillivray RT, Di Cera E. Determinants of specificity in coagulation proteases. J Thromb Haemost. 2005;3:2401–2408. doi: 10.1111/j.1538-7836.2005.01456.x. [DOI] [PubMed] [Google Scholar]
- 3.Esmon CT. Regulation of blood coagulation. Biochim Biophys Acta. 2000;1477:349–360. doi: 10.1016/s0167-4838(99)00266-6. [DOI] [PubMed] [Google Scholar]
- 4.Gohara DW, Di Cera E. Allostery in trypsin-like proteases suggests new therapeutic strategies. Trends Biotechnol. 2011;29:577–585. doi: 10.1016/j.tibtech.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Cera E. Thrombin as an anticoagulant. Prog Mol Biol Transl Sci. 2011;99:145–184. doi: 10.1016/B978-0-12-385504-6.00004-X. [DOI] [PubMed] [Google Scholar]
- 6.Lechtenberg BC, Freund SM, Huntington JA. An ensemble view of thrombin allostery. Biol Chem. 2012;393:889–898. doi: 10.1515/hsz-2012-0178. [DOI] [PubMed] [Google Scholar]
- 7.Adams TE, Huntington JA. Thrombin-cofactor interactions: structural insights into regulatory mechanisms. Arterioscler Thromb Vasc Biol. 2006;26:1738–1745. doi: 10.1161/01.ATV.0000228844.65168.d1. [DOI] [PubMed] [Google Scholar]
- 8.Malovichko MV, Sabo TM, Maurer MC. Ligand binding to anion-binding exosites regulates conformational properties of thrombin. J Biol Chem. 2013;288:8667–8678. doi: 10.1074/jbc.M112.410829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nussinov R, Tsai CJ. Unraveling structural mechanisms of allosteric drug action. Trends Pharmacol Sci. 2014;35:256–264. doi: 10.1016/j.tips.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Tsai CJ, Nussinov R. A unified view of “how allostery works”. PLoS Comput Biol. 2014;10:e1003394. doi: 10.1371/journal.pcbi.1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sidhu PS, Liang A, Mehta AY, Abdel Aziz MH, Zhou Q, Desai UR. Rational design of potent, small, synthetic allosteric inhibitors of thrombin. J Med Chem. 2011;54:5522–5531. doi: 10.1021/jm2005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehta AY, Thakkar JN, Mohammed BM, Martin EJ, Brophy DF, Kishimoto T, Desai UR. Targeting the GPIbalpha binding site of thrombin to simultaneously induce dual anticoagulant and antiplatelet effects. J Med Chem. 2014;57:3030–3039. doi: 10.1021/jm4020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henry BL, Monien BH, Bock PE, Desai UR. A novel allosteric pathway of thrombin inhibition: Exosite II mediated potent inhibition of thrombin by chemo-enzymatic, sulfated dehydropolymers of 4-hydroxycinnamic acids. J Biol Chem. 2007;282:31891–31899. doi: 10.1074/jbc.M704257200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olson ST, Swanson R, Raub-Segall E, Bedsted T, Sadri M, Petitou M, Herault JP, Herbert JM, Bjork I. Accelerating ability of synthetic oligosaccharides on antithrombin inhibition of proteinases of the clotting and fibrinolytic systems. Comparison with heparin and low-molecular-weight heparin. Thromb Haemost. 2004;92:929–939. doi: 10.1160/TH04-06-0384. [DOI] [PubMed] [Google Scholar]
- 15.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3. Springer; New York: 2006. Quenching of Fluorescence; pp. 278–330. [Google Scholar]
- 16.Olson ST, Halvorson HR, Bjork I. Quantitative characterization of the thrombin-heparin interaction. Discrimination between specific and nonspecific binding models. J Biol Chem. 1991;266:6342–6352. [PubMed] [Google Scholar]
- 17.Record MT, Jr, Lohman ML, De Haseth P. Ion effects on ligand-nucleic acid interactions. J Mol Biol. 1976;107:145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- 18.Desai UR, Petitou M, Bjork I, Olson ST. Mechanism of heparin activation of antithrombin. Role of individual residues of the pentasaccharide activating sequence in the recognition of native and activated states of antithrombin. J Biol Chem. 1998;273:7478–7487. doi: 10.1074/jbc.273.13.7478. [DOI] [PubMed] [Google Scholar]
- 19.Vindigni A, White CE, Komives EA, Di Cera E. Energetics of thrombin-thrombomodulin interaction. Biochemistry. 1997;36:6674–6681. doi: 10.1021/bi962766a. [DOI] [PubMed] [Google Scholar]
- 20.Li CQ, Vindigni A, Sadler JE, Wardell MR. Platelet glycoprotein Ib alpha binds to thrombin anion-binding exosite II inducing allosteric changes in the activity of thrombin. J Biol Chem. 2001;276:6161–6168. doi: 10.1074/jbc.M004164200. [DOI] [PubMed] [Google Scholar]
- 21.De Cristofaro R, De Candia E, Rutella S, Weitz JI. The Asp(272)-Glu(282) region of platelet glycoprotein Ibalpha interacts with the heparin-binding site of alpha-thrombin and protects the enzyme from the heparin-catalyzed inhibition by antithrombin III. J Biol Chem. 2000;275:3887–3895. doi: 10.1074/jbc.275.6.3887. [DOI] [PubMed] [Google Scholar]
- 22.Lechtenberg BC, Freund SM, Huntington JA. GpIbalpha Interacts Exclusively with Exosite II of Thrombin. J Mol Biol. 2014;426:881–893. doi: 10.1016/j.jmb.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zarpellon A, Celikel R, Roberts JR, McClintock RA, Mendolicchio GL, Moore KL, Jing H, Varughese KI, Ruggeri ZM. Binding of alpha-thrombin to surface-anchored platelet glycoprotein Ib(alpha) sulfotyrosines through a two-site mechanism involving exosite I. Proc Natl Acad Sci U S A. 2011;108:8628–8633. doi: 10.1073/pnas.1017042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jandrot-Perrus M, Clemetson KJ, Huisse MG, Guillin MC. Thrombin interaction with platelet glycoprotein Ib: effect of glycocalicin on thrombin specificity. Blood. 1992;80:2781–2786. [PubMed] [Google Scholar]
- 25.Sidhu PS, Abdel Aziz MH, Sarkar A, Mehta AY, Zhou Q, Desai UR. Designing allosteric regulators of thrombin. Exosite 2 features multiple subsites that can be targeted by sulfated small molecules for inducing inhibition. J Med Chem. 2013;56:5059–5070. doi: 10.1021/jm400369q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdel Aziz MH, Mosier PD, Desai UR. Identification of the site of binding of sulfated, low molecular weight lignins on thrombin. Biochem Biophys Res Commun. 2011;413:348–352. doi: 10.1016/j.bbrc.2011.08.102. [DOI] [PMC free article] [PubMed] [Google Scholar]


