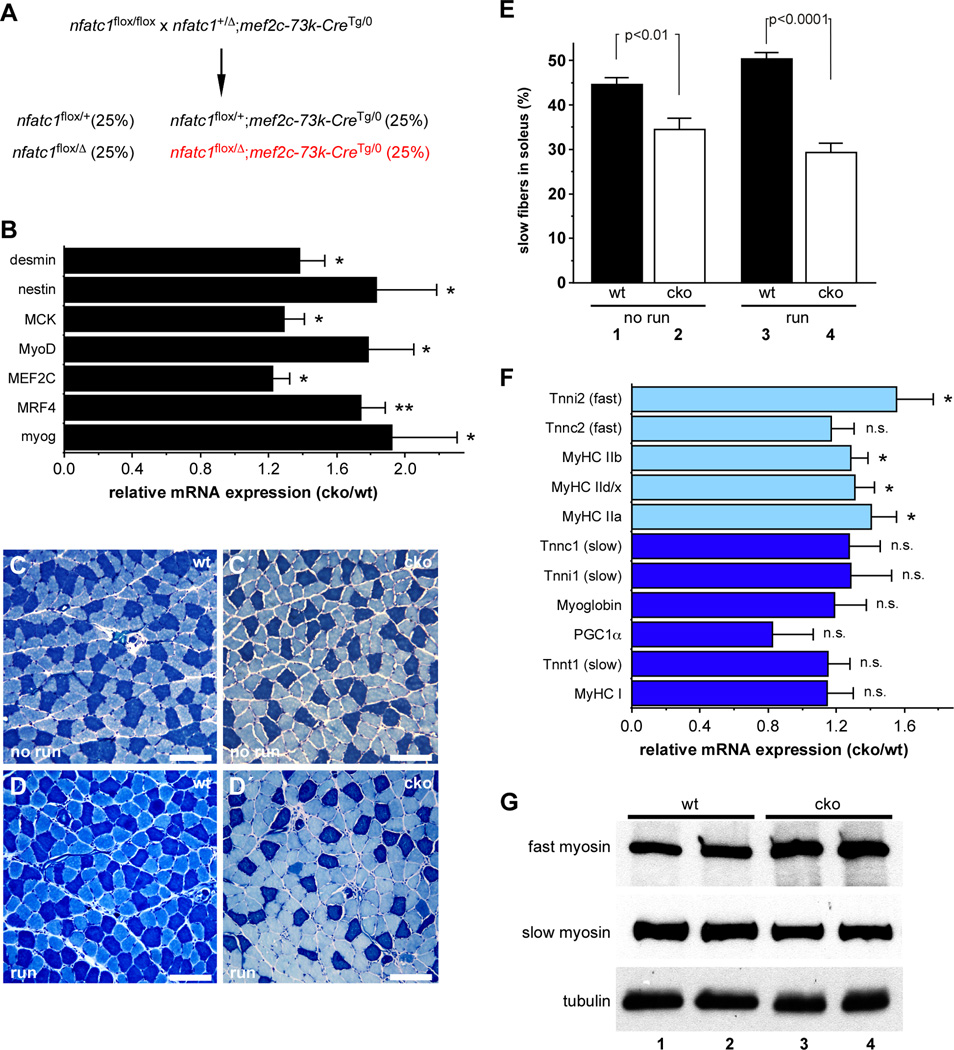
Figure 2. NFATc1 is required for normal fiber type composition, gene expression, and exercise-induced fiber type switching in vivo.
(A) Strategy used to generate skeletal muscle specific knockout of nfatc1. (B) qPCR analysis of MyoD target gene expression in the soleus muscles of adult control (wt) and nfatc1SkMKO (cko) mice. *, p < 0.05; **, p < 0.01. Data shown represent the mean ratio of expression in nfatc1SkMKO compared to wild type muscle plus SEM for 6 mice of each genotype. (C–E) Metachromatic ATPase staining of soleus muscles showed a higher percentage of slow fibers (dark blue) in control (wt, panels C, D) than in nfatc1SkMKO (cko, panel C′, D′) in the absence of exercise (no run; C, C′) or following 7 days of voluntary exercise (run; D, D′). Scale bars =100µm. (E) No run wt mice had a significantly higher percentage of slow fibers than unexercised nfatc1SkMKO mice (compare lanes 1 and 2, p < 0.01). Following exercise, the percentage of slow fibers in wt soleus increased from 45% to 50% (compare lanes 1 and 3, p < 0.05); the percentage of slow fibers in the soleus of nfatc1SkMKO mice showed no statistically significant difference (compare lanes 2 and 4). Data are presented as the mean percentage of slow fibers plus SEM for 6 mice in each group. (F) qPCR analysis of fast (light blue bars) and slow (dark blue bars) fiber gene expression in the soleus muscles of unexercised control (wt) and nfatc1SkMKO (cko) male mice. Data are shown as the mean ratio of expression in nfatc1SkMKO to wild type muscle plus SEM for 6 mice of each genotype. *, p < 0.05; n.s., not significant. (G) Western blot analysis of fast and slow myosin expression from wt (lanes 1, 2) and nfatc1SkMKO (cko; lanes 3, 4) soleus muscles. α-tubulin was examined as a loading control on the same cell lysates. See also Figure S2 and Figure S3.