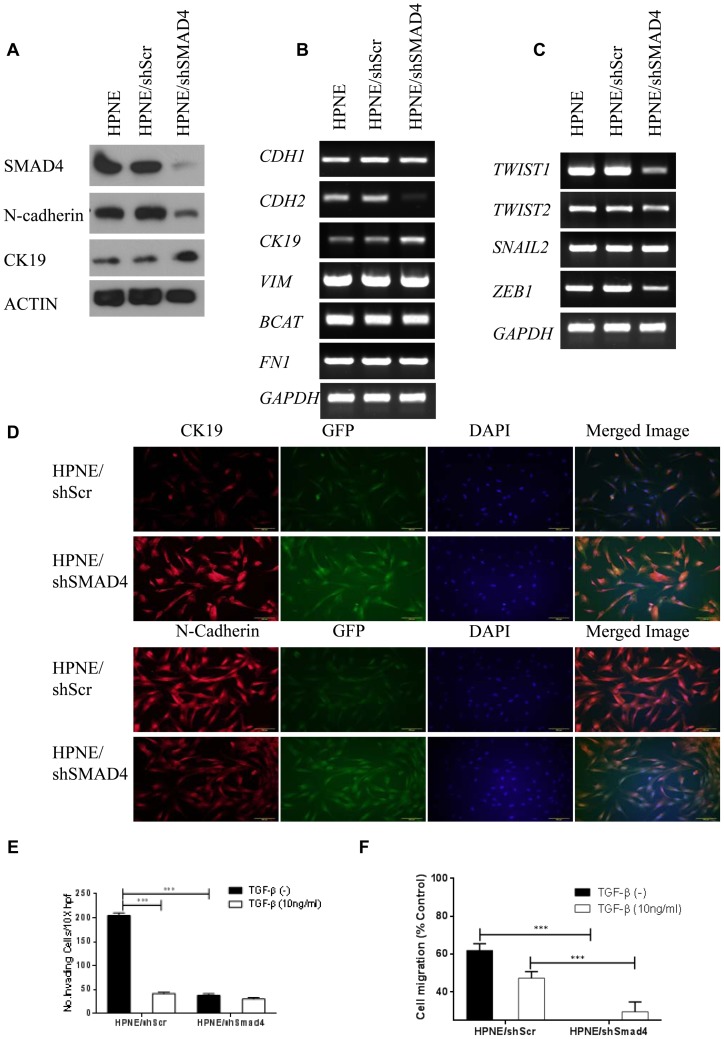
Figure 1. Knocked down SMAD4 reduces N-cadherin protein level and inhibits invasion and migration in HPNE cells.
(a) Western blot analysis of cell expression of SMAD4 and N-cadherin and CK19. Actin was used as the loading control. (b) RT-PCR results showing cell mRNA levels of CDH1, CDH2, CK19, VIM, BCAT, and FN1, and (c) TWIST1, TWIST2, SNAIL2, and ZEB1. GAPDH was used as the housekeeping control. (d) Immunofluorescence staining of CK19 and N-cadherin in HPNE/shScr or HPNE/shSMAD4 cells. CK19 and N-cadherin were labeled with red fluorescent Alexa Fluor 594 goat anti-rabbit IgG (A11012, Invitrogen). GFP-positive cells represent cells transfected with shScr or shSMAD4. Nuclei were counterstained with blue fluorescent 4,6-diamidino-2-phenylindole. Images were merged using Olympus CellSens software. (e) Modified Boyden chamber assay. HPNE/shScr and HPNE/shSMAD4 Cells were added with or without 10 ng/ml TGF-β to serum-free media inserts in the top chamber, and 20% FBS was placed in the bottom chamber as a chemoattractant. Invasive cells were counted in 3 fields at 10× magnification in duplicated inserts. (f) Wound-healing assay. HPNE/shScr and HPNE/shSMAD4 cells were treated with or without10 ng/ml TGF-β. The y-axis represents cell migration distance at the time of the scratch and after 20 hours. Three random images (4×) were taken at these time points, and migration rate was determined as the ratio of distance at 20 hours versus 0 hours in the wound's gap using Adobe Photoshop software. Results are the mean ± s.d. of 3 independent experiments.