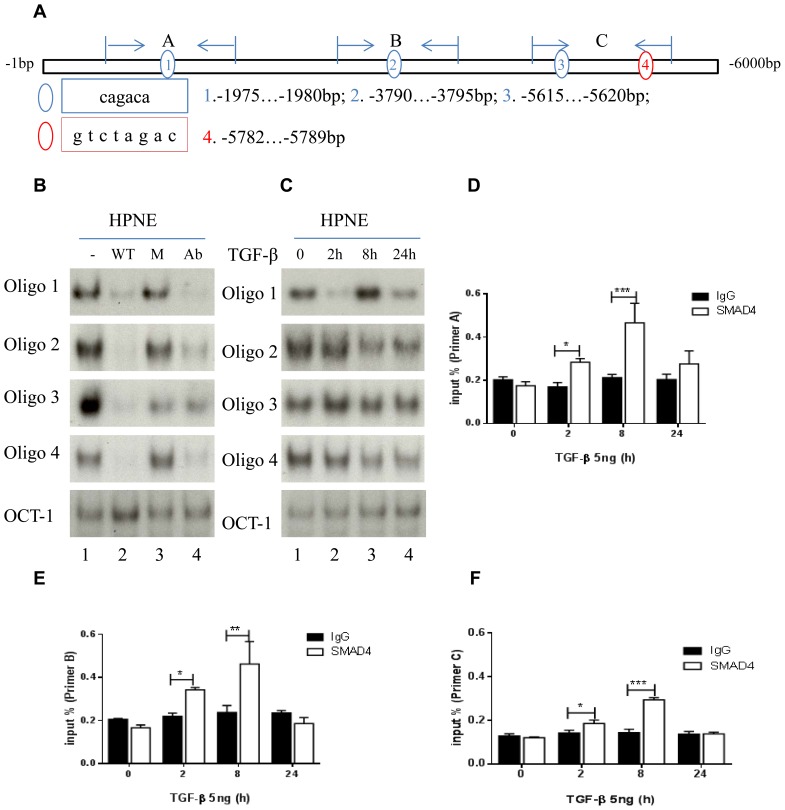
Figure 3. Map of multiple SBEs in CDH2 promoter.
(a) Three SBEs with a CAGACA sequence (blue circles 1, 2, and 3) and one SBE with a GTCTAGAC sequence (red circle 4). Sections A, B, and C represent 3 primers and an amplifying region for ChIP assay in the promoter. (b) Electrophoretic mobility shift assay results showed that 4 SBE oligos had strong DNA and nuclear protein interaction bands (lane 1), binding was quenched by wild-type (WT) oligos (lane 2) and not by mutant (M) oligos (lane 3), and anti-SMAD4 antibody (Ab) inhibited binding activity (lane 4). (c) SBE binding activity was regulated by TGF-β treatment at 0, 2, 8, and 24 hours. Oct-1 DNA binding activities were determined as loading controls in (b) and (c). (d-f) ChIP assays and real-time PCR of primers A, B, and C comparing the ratio of IgG to anti-SMAD4 antibody with or without 5 ng of TGF-β at 0 2, 8, and 24 hours. *P<0.05, **P<0.01, ***P<0.001.