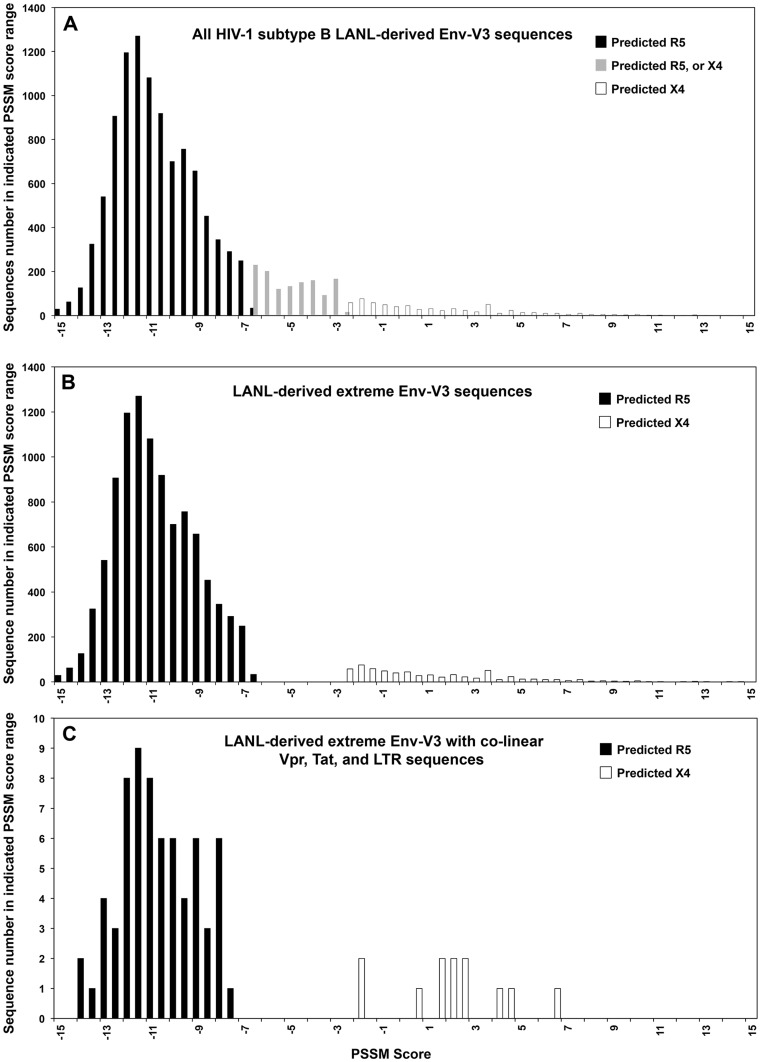
Figure 1. Quantitative PSSM score analysis of LANL-derived Env-V3 sequences.
(A) A total of 11,866 HIV-1 subtype B Env-V3 sequences with a complete 35-amino-acid sequence were retrieved from the LANL database. PSSM scores were obtained and results were plotted. Black columns represent predicted R5 viral sequences; white columns represent predicted X4 viral sequences; and gray columns represent an area of mixture of Env-V3 sequences, which were predicted as either X4 or R5. (B) A total of 10,600 Env-V3 sequences that contained 35-amino-acid residues with PSSM scores below −6.96 and therefore classified as R5 (black column) with scores above −2.88 classified as X4 (white column). The gray column area was eliminated from the subsequent analysis. (C) Only the final 79 LANL-derived Env-V3 sequences were included in this study. Of these, 67 sequences were predicted to utilize CRR5, while 12 sequences were predicted to be CXCR4 utilizing. The frequencies of PSSM scores were analyzed and the distributions were compared between the two groups.