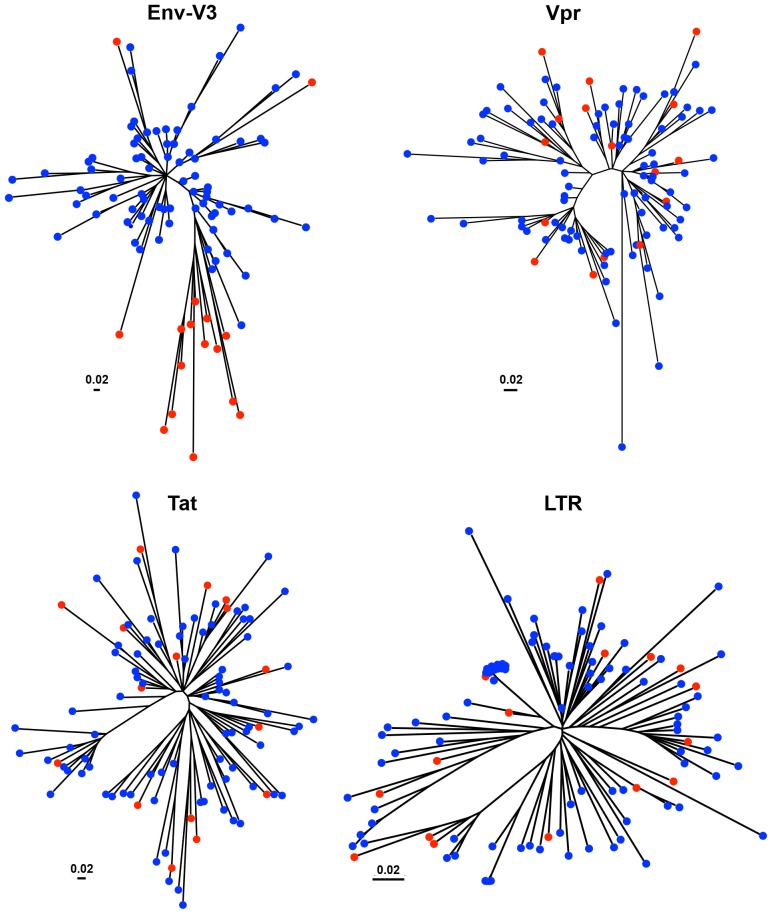
Figure 4. Inferred neighbor-joining phylogenetic tree of HIV-1 Env-V3 sequences derived from LANL and Drexel Medicine (DM).
The phylogenetic trees of Env-V3, Vpr, Tat, and LTR sequences were inferred by utilizing the maximum-likelihood method using the JTT with frequency model. The trees with the highest log likelihood are shown. A total of LTR sequences were applied in an evolution tree construction, which was based on the maximum-likelihood accompanied with data-specific model, and the highest log likelihood is presented. Initial tree(s) for the heuristic search were obtained automatically as follows. The trees were drawn to scale, with branch lengths measured in the number of substitutions per site. Tree analysis was conducted in MEGA5 and based on a total of 103 sequences in each tree. The predicted X4 sequences are indicated with red circles, and the blue circles represent the predicted R5 sequence.