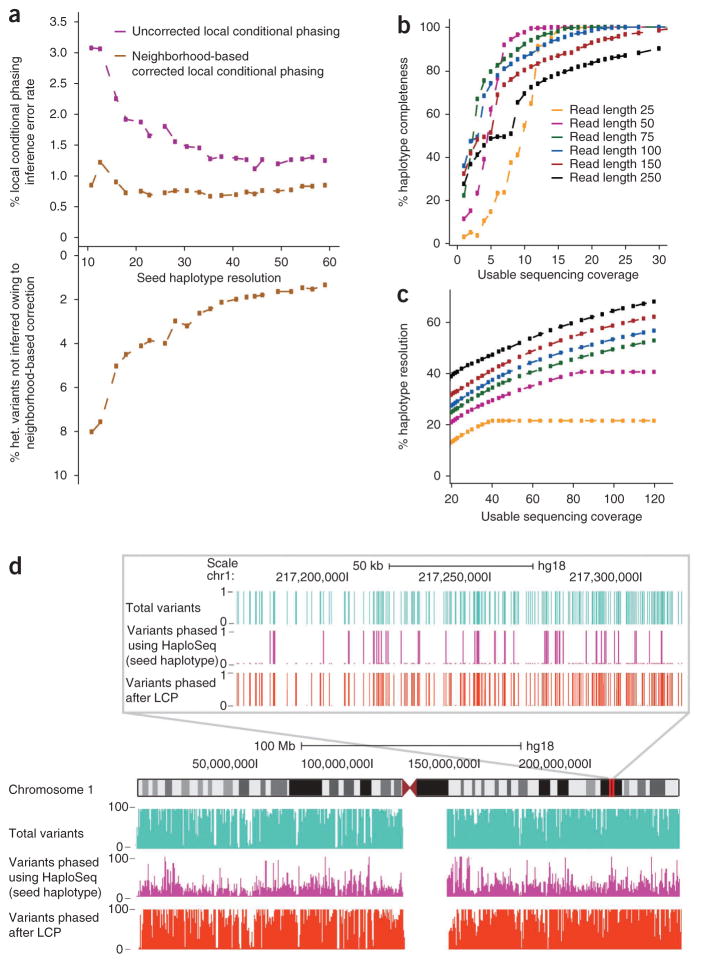
Figure 5.
HaploSeq analysis coupled with local conditional phasing permits high-resolution haplotype reconstruction in humans. (a) Local conditional phasing. The x axis is the chromosome span seed haplotypes resolution generated by simulation. The top panel shows the error rates of local conditional phasing using both an uncorrected (purple) and neighborhood corrected phasing (gold, window size = 3). Because of neighborhood correction, some variants cannot be locally inferred. The bottom panel shows the percentage of variants that remain unphased due to neighborhood correction as a function of resolution. All simulations are done using GM12878 chromosome 1. (b) Chromosome-spanning seed haplotype (MVP block) at varying parameters of read length and coverage. All simulations are done in GM12878 chromosome 1. (c) Different combinations of read length and coverage generate high-resolution seed haplotypes. Resolution metric depends on percentage of completeness. For example, for 250 bp reads at 30× coverage, resolution is 45% of the 90% variants spanned in haplotype. All simulations are done in GM12878 chromosome 1. (d) UCSC genome browser shot analogous to Figure 4b, illustrating phasing by local conditional phasing (LCP).