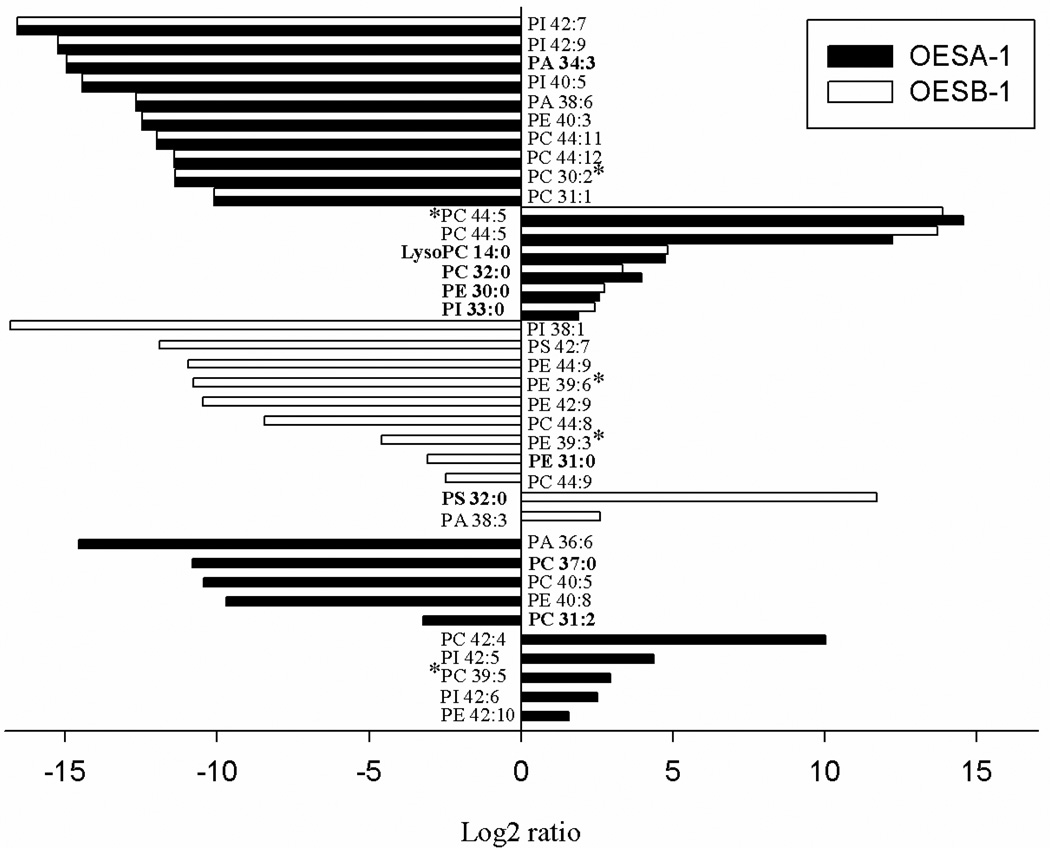
Fig. 6. Changes in phospholipid abundance in sPLA2 overexpressing strains.
Summary of the phospholipids, whose levels were found to be consistently altered in OESA-1 (solid bars) and OESB-1 (open bars) compared to the control (NGA142-1) strain. Phospholipids that changed by at least two-fold in the overexpressing strains are reported; changes are annotated as the log2 ratio of the amount of each phospholipid in OESA-1 and OESB-1 relative to the control. Individual phospholipid and lysophospholipid species matching the available yeast lipid template are in bold, the remaining species were identified against a animal lipid template; lipid species that did not match neither the yeast nor the animal lipid template, were identified on the basis of their spectra and m/z values and are indicated with an asterisk. PI, phosphatidylinositol; PA, phosphatidic acid; PE, phosphatidylethanolamine; PC, phosphatidylcholine. Numbers are the length of the carbon chain: degree of unsaturation.