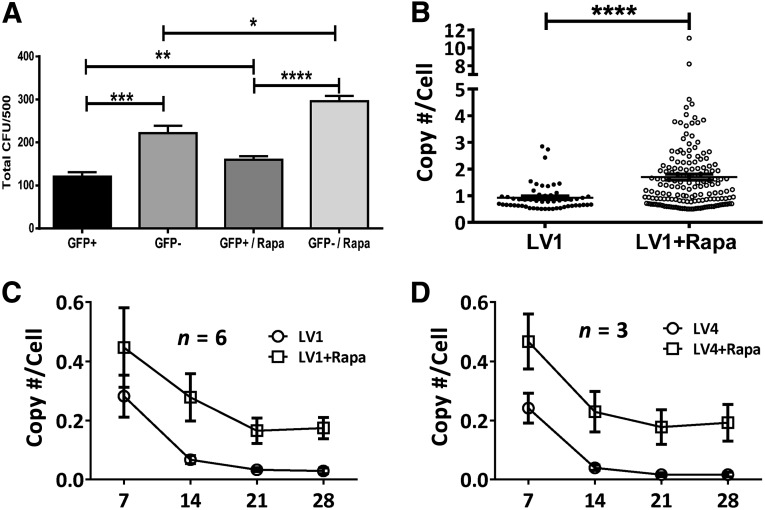
Figure 4.
Analysis of colony-forming potential and transgene copy number following transduction in rapamycin. (A): Rapamycin enhanced colony-forming potential of genetically modified cells. CD34+ cells were transduced with LV1 (multiplicity of infection, 10) after overnight prestimulation in SFT6 followed by 7 days of expansion in SFEM SFT6 SR1. GFP+ and GFP− cells were sorted and plated for CFU. Statistical analysis was performed with paired t test (n = 3). (B): Methylcellulose colonies of LV1-transduced cells with or without rapamycin treatment were picked, lysed, and analyzed by quantitative polymerase chain reaction with WPRE- and apolipoprotein B (ApoB)-specific primers as described in Materials and Methods. The ratio of transgene copy number determined by WPRE and cell number determined by ApoB were calculated and presented. The data sets were analyzed for statistical significance by paired t test. (C, D): Stability of transgene average copy number per cell in 4 weeks differentiation cultures of transduced hematopoietic stem and progenitor cells using LV1 (C) and LV4 (D). Height of bars represents the average value, and error bars represent SEM. ∗, p < .05; ∗∗, p < .01; ∗∗∗, p < .001; and ∗∗∗∗, p < .0001. Abbreviations: CFU, colony-forming unit; GFP, green fluorescent protein; LV, lentivirus; Rapa, rapamycin; WPRE, woodchuck hepatitis virus post-translational regulatory element.