Figure 4.
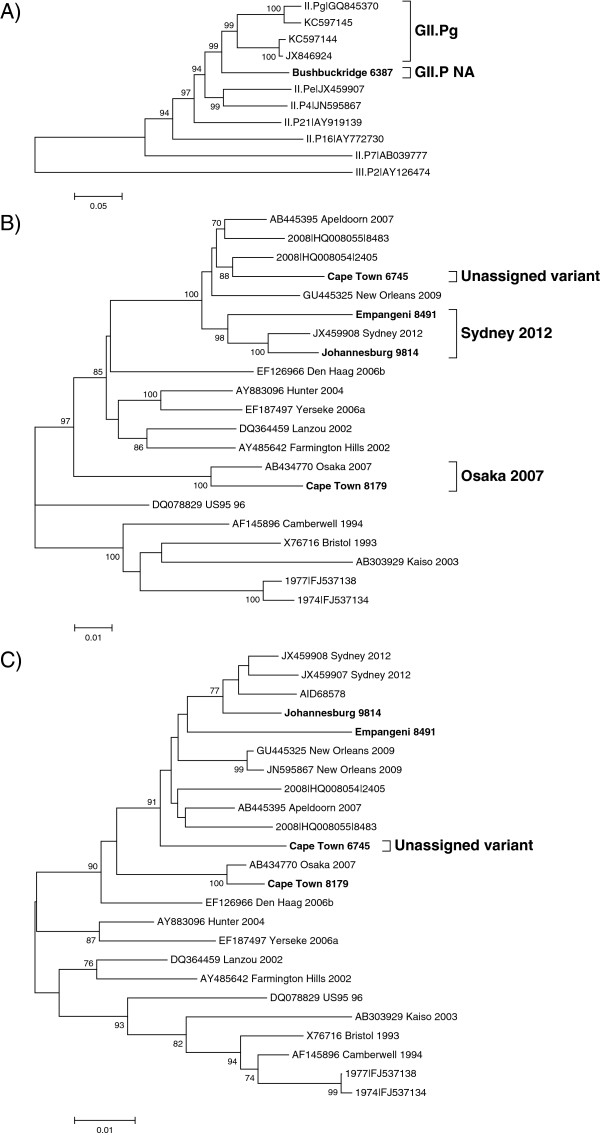
Phylogenetic analysis of unassigned norovirus (NoV) recombinants. Neighbor-joining phylogenetic analysis of A) the complete polymerase gene (1533 bp) of Bushbuckridge 6387 (GII.P NA/GII.3), B) the complete capsid (1623 bp) gene of NoV GII.4 variants and, C) the 540 amino acid sequence encoded by the capsid gene of the NoV GII.4 variants. For the amino acid tree the evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. The NoV reference strains are indicated by GenBank accession numbers as well as GII.4 variant names and the strains identified in this study are shown in bold. Statistical significance was evaluated with a 1000 bootstrap replicates and bootstrap support of >70% is indicated. The scale bar represents nucleotide or amino acid substitutions per site.
