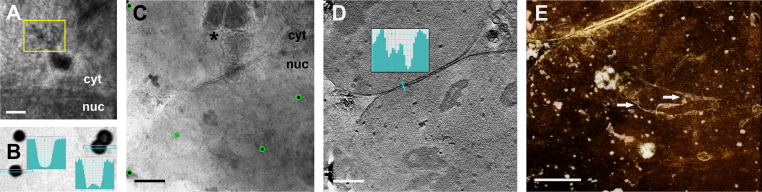
Fig. 1.
Quantum dot-containing nanoparticles as alignment markers in cryoXT of vitreous mammalian cells. (A) Sub-area of the zero degree image taken from a tilt series of a nucleus of a rabbit kidney epithelial-like RK13 cell (sample thickness: 8 µm) stably expressing LBR1TM-GFP (cf. Supplement Movie 1, pre-aligned tilt series). Note the low absorbance contrast of photoluminescent aqueous CdSe/ZnS microspheres (λmax.em.=625 nm; diameter: ~150 nm; yellow frame) as compared to standard gold-coated silica beads (center). (B) Density profiles of the other tested multimodal nanoparticles, polyelectrolyte-Qdot® 605 coated gold beads (left; diameter: 208 nm) and standard gold-coated silica beads (right; diameter: 267 nm) in a zero degree image of a cryoXT tilt series (sample thickness: 3 µm). Note that the polyelectrolyte-Qdot® 605 coat (λmax.em.=605 nm) of the former nanoparticles, although imaged without cellular background in a hole of the Quantifoil carbon coat of the grid, did not provide sufficient contrast to be detected separately from the gold core (total diameter of the nanoparticles measured by dynamic light scattering: 277 nm, standard deviation 8 nm; cf. Supporting information). (C–E) Soft X-ray cryo-tomography of two nuclei before cytokinesis in a RK13 cell expressing LBR1TM-GFP, incubated on-grid with 20 µM Saquinavir for 48 h. The tilt series was aligned with the help of polyelectrolyte-Qdot® 605 coated gold beads as alignment markers (green circles in C: three polyelectrolyte-Qdot® 605 coated gold beads in the periphery of the image frame, and two cellular marker points, most probably lipid bodies, were used for alignment; asterisk: ice contamination on the sample surfaces; cf. Supplement Movie 2, pre-aligned tilt series). The next panels show the corresponding reconstruction (D, cyan bar: measuring position of the density profile shown as inset; profile determined from unbinned data, pixel size: 9.9 nm, revealing the inner and outer nuclear membrane of the upper nucleus with a distance of 40 nm; cf. Supplement Movie 3, sliced view through the tomographic reconstruction; thickness of reconstructed volume: 4.1 µm) and visualization as rendered volume with inverse contrast (E, arrows: nucleoplasmic reticulum; Supplement Movie 4, animated sub-volume of the lower nucleus). Scale bars are 500 nm (A) and 2 µm (C–E). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)