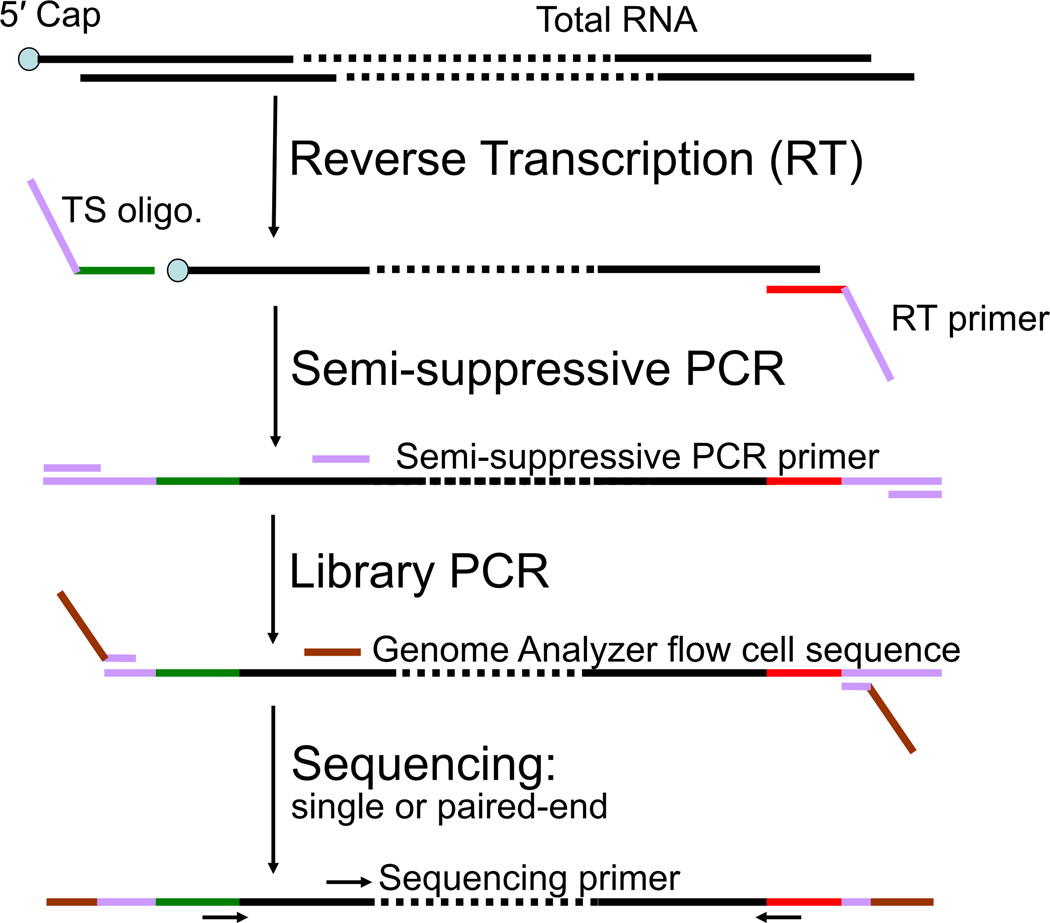
Figure 1.
Flowchart of nanoCAGE protocol. Briefly, the template switching (TS) oligonucleotide is added to the first-strand cDNA synthesis reaction along with the reverse transcription (RT) primer. The three guanosine ribonucleosides at the 3′ end of the template switching primer hybridize to cytidine deoxynucleosides added to the 3′ end of the newly synthesized cDNA strand by the reverse transcriptase in a cap-dependent manner (Hirzmann et al., 1993). After hybridization, the reverse transcriptase will extend the cDNA strand using the template switching oligonucleotide as a template. Hence, the cDNA originating from a capped RNA will have at its 3′ end sequence reverse-complementary to the sequence of the template switching oligonucleotide. These sequences are needed for the synthesis and amplification of the second cDNA strand by semi suppressive PCR, that minimizes the amplification of shorter artifacts like primer dimers or aberrant cDNAs template-switched from a RT primer or reverse-transcribed from a TS oligonucleotide. End-sequences required for sequencing the cDNAs on the Genome Analyzer II are introduced by PCR (“Library PCR”), and the cDNAs are then sequenced as single or paired ends.