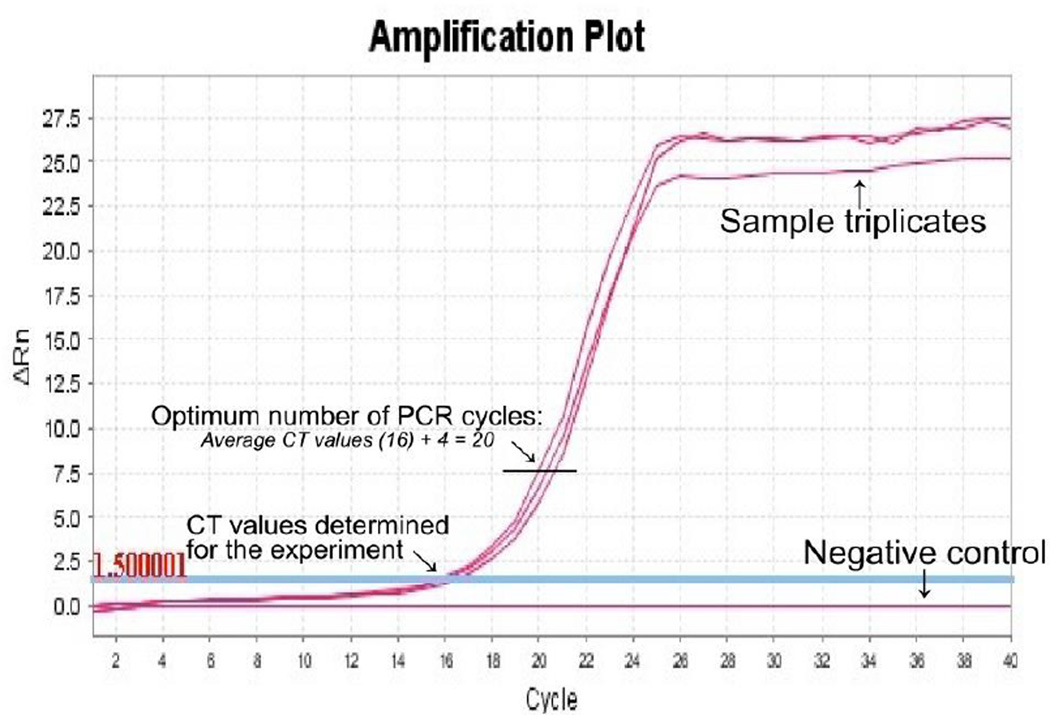
Figure 3.
An example for the determination of the optimum number of PCR cycles for the synthesis of second strand cDNAs by semi-suppressive PCR. A cycle threshold (CT) value is determined for the experiment done by quantitative real-time PCR. The CT value is automatically defined by the qPCR software as the number of cycles required for the fluorescent signal to cross the threshold i.e., to exceeds background level. The optimum cycle number for large scale synthesis of the second strand cDNAs can be determined by adding 4 cycles to the detected CT.