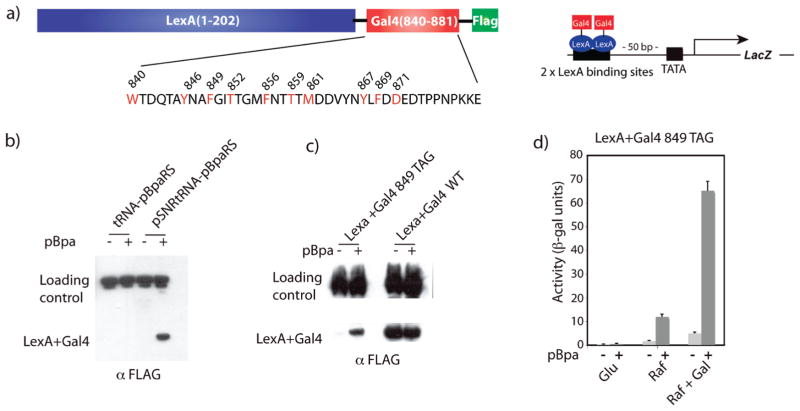
Figure 2.
In vivo incorporation of pBpa into FLAG-tagged LexA+Gal4 Phe849pBpa. (a) To assess the impact of pBpa on the function and binding modes of the Gal4 TAD, plasmids encoding the DNA binding domain of LexA fused to Gal4(840–881) as well as a FLAG tag were constructed. The LexA DBD was utilized to exclusively examine transcriptional activation at the two unique LexA binding sites upstream of the LacZ reporter. Positions at which pBpa mutagenesis was carried out are indicated in red. (b) Yeast cells bearing plasmids encoding LexA+Gal4 849 TAG and the pBpa-specific tRNA/synthetase pair expressed by ptRNA-pBpaRS or pSNRtRNA-pBpaRS were grown in the presence or absence of 2 mM pBpa and analyzed by Western blot (α-FLAG). (c) Yeast cells bearing plasmids encoding LexA+Gal4 849 TAG or LexA+Gal4 WT and the pBpa-specific tRNA/synthetase pair pSNRtRNA-pBpaRS were analyzed as described in (b). (d) β-Galactosidase assays were used to measure the activity of the LexA+Gal4 849 TAG mutant in the presence or absence of 2 mM pBpa under permissive or restrictive conditions. Each activity is the average of values from at least three independent experiments with the indicated error (SDOM). See Supporting Information for additional details.