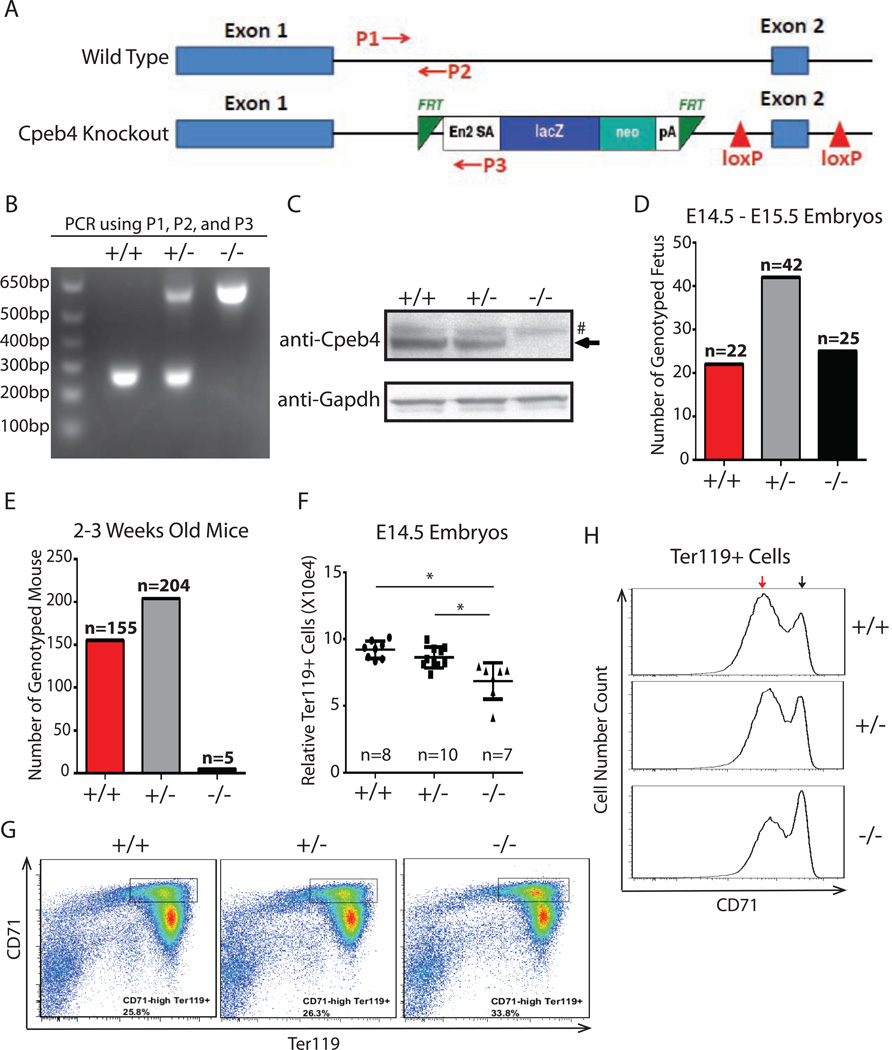
Figure 7. Cpeb4 is functionally important in vivo.
(A) Structures of Cpeb4 genomic region in wild type and Cpeb4 knockout mouse. P1, P2, and P3 are PCR primers for genotyping. (B) Genotyping of wildtype (+/+), Cpeb4 heterozygous (+/−), and Cpeb4 knockout (−/−) mice by PCR. (C) Western blot of E14.5 total fetal liver cells isolated from wildtype, Cpeb4 heterozygous, and Cpeb4 homozygous knockout embryos. (D) Genotyping of E14.5-E15.5 embryos from a mating of Cpeb4 heterozygotes. (E) Genotyping of 2–3 week old mice from matings of Cpeb4 heterozygotes. (F) The quantification of Ter119+ cells in fetal livers from E14.5 embryos. The y axle is the scale relative to a known amount of added beads. * indicates p < 0.05 using two-tailed Student’s t test. (G) CD71 and Ter119 staining of total E14.5 fetal liver cells. (H) CD71 expression levels in Ter119+ cells from E14.5 mouse fetal liver. The black arrow indicates high CD71 level; the red arrow indicates low CD71 level. The results of (G, H) are representative of E14.5 embryos from 4 sets of matings of Cpeb4 heterozygotes.