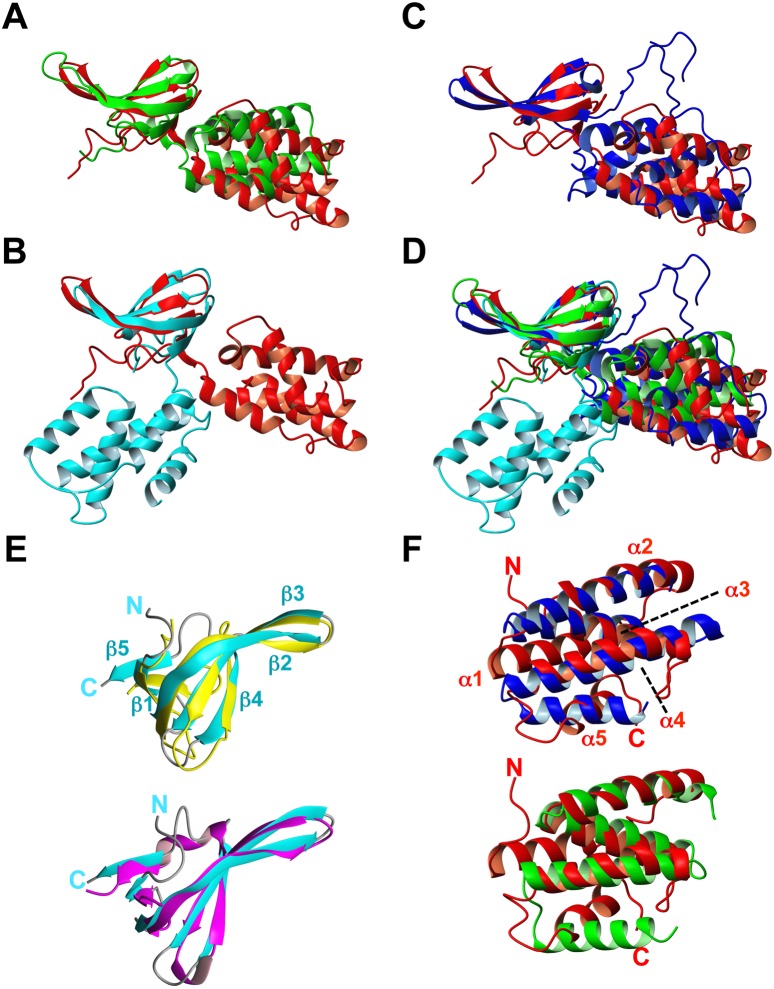
Figure 3. Structural comparisons with M. xanthus CdnL NMR structure.
Backbone overlay of the CdnL NMR structure (red) onto the crystal structures of (A): TtCdnL (green; PDB: 4L5G); (B) one unit in the MtCdnL domain-swapped dimer (blue; PDB: 4ILU); (C) MtCdnL in complex with the RNAP β-lobe domain (cyan; PDB: 4KBM); (D) all four structures. The overlay shown was generated with optimal (maximum) superposition of the N-terminal domains of each structure. (E) Backbone overlay of CdnLNt (cyan; left) onto: (top) TtCdnLNt (gold; PDB ID: 2lqK); (bottom) E. coli TRCF-RID (magenta; PDB ID: 2eyq). (F) Backbone overlay of CdnLCt (red) onto the TPR domains of: (top) FK506-binding protein (blue; PDB ID: 1kt0); (bottom) CadCpd (green; PDB ID: 3ly8). Structures were generated with MOLMOL.