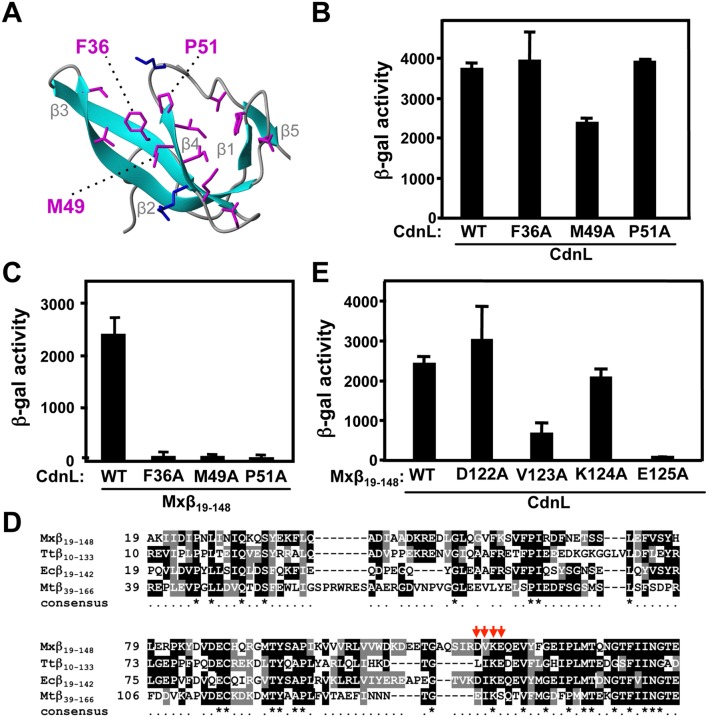
Figure 4. Mutational analysis of CdnL interaction with RNAP-β.
(A) Native CdnLNt structure showing residue side chains that may contact RNAP-β, with the three labeled tested by mutational analysis (see text). (B) BACTH analysis showing that the F36A, M49A and P51A CdnL variants self-interact in cells with the pKT25 and pUT18 constructs of wild type or mutant cdnL, as indicated. (C) BACTH analysis of the interaction with Mxβ19–148 (in pUT18C) of CdnL F36A, M49A, or P51A mutants in pKT25. (D) Sequence alignment of the M. xanthus RNAP-β segment, Mxβ19–148 (NCBI code, YP_631280.1) and its equivalents in T. thermophilus, Ttβ10–133 (WP_014630291), in E. coli, Ecβ19–142 (P0A8V2), and in M. tuberculosis (CAB09390). Residues shaded black if identical, or grey if similar, in at least two of the aligned sequences. An asterisk in the consensus line below indicates conservation in all four sequences. Red arrows point to RNAP-β residues analyzed by site-directed mutagenesis in this study. (E) BACTH analysis of the interaction of Mxβ19–148 (WT) or its indicated mutants in pUT18C versus wild-type CdnL in pKT25.