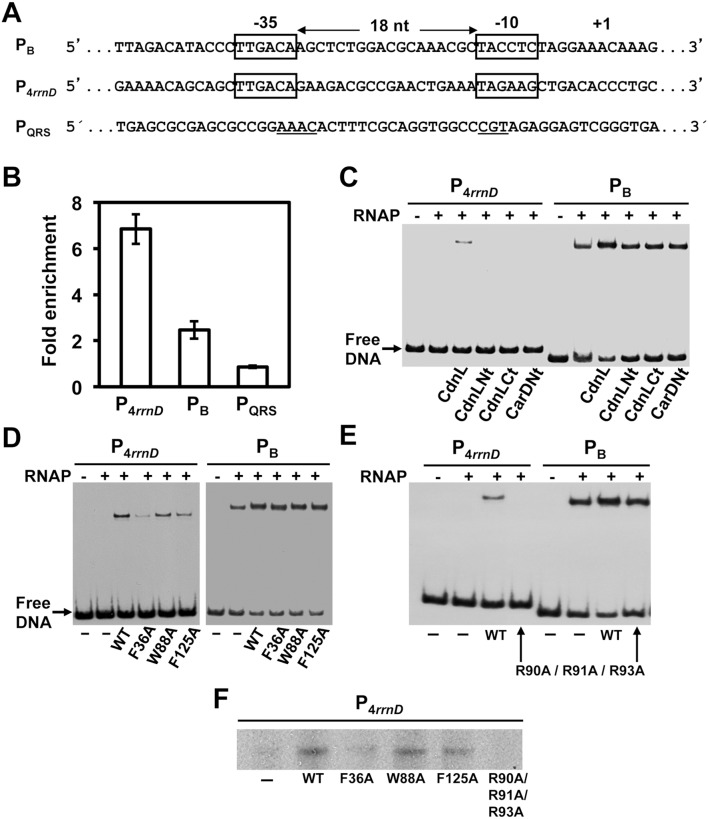
Figure 7. CdnL favors RPo formation and transcription from σA-dependent promoters.
(A) Promoter sequences for the σA-dependent P4rrnD and PB, indicating the -35 and -10 elements (in boxes) and the transcription start site. For comparison, the ECF-σ CarQ-dependent PQRS promoter is also included, with the conserved motifs at the -35 and -10 regions underlined. (B) ChIP-qPCR analysis of CdnL enrichment at P4rrnD, PB and PQRS in M. xanthus carried out as described in the text. The mean and standard of three independent experiments are shown. (C)–(E) EMSA of the binding of the 32P-labeled 130-bp PB or 151-bp P4rrnD DNA probes to 130 nM RNAP-σA holoenzyme, alone or in the presence of 10 µM of CdnL, CdnLNt, CdnLCt, or CarDNt (C), or a given CdnL variant (D, E), as indicated. After incubation for 30-min at 37°C, the complexes formed were challenged by adding 1 µg heparin. Note that any alteration in the migration of the shifted complexes due to CdnL binding to RNAP was not discernible, possibly because CdnL is considerably smaller than RNAP. (F) Single round, run-off in vitro transcription from P4rrnD (see Materials and methods) with 130 nM RNAP-σA alone or with 10 µM CdnL (WT or mutant) followed by heparin challenge (1 µg) and then addition of the labeled NTP mix to initiate transcription. In C–F, representative data from three or more experiments are shown.