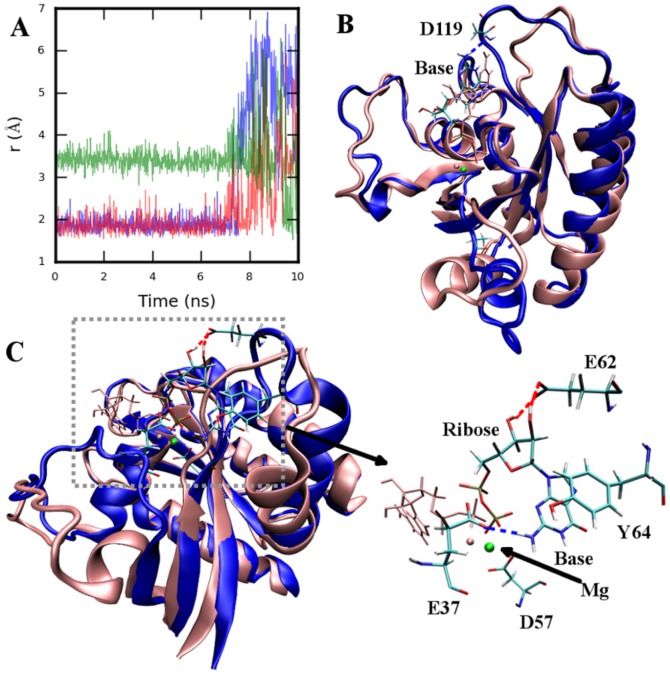
Figure 4. All-atom SMD and CMD of 4Q21-OpenSI at 360K.
A. Distance between atom pairs D119OD1-baseH1 (blue), D119OD2-baseH21 (red), D119OD2-baseH22 (green). B. Structure at the end of SMD simulation (blue) superimposed on top of starting structure (pink). Base is displaced from its starting position (GDP drawn in pink) as D119 is pulled. C. Structure at the end of the CMD simulation (blue) continued from the last snapshot of SMD. Starting position of GDP/Mg is shown in pink. Both GDP/Mg is destabilized and displaced away from the nucleotide pocket. E37, D57, E62, and Y64 are shown stabilizing the leaving GDP/Mg. Structure alignment was performed using backbone atoms N, CA, and C of core residues as defined in [18].