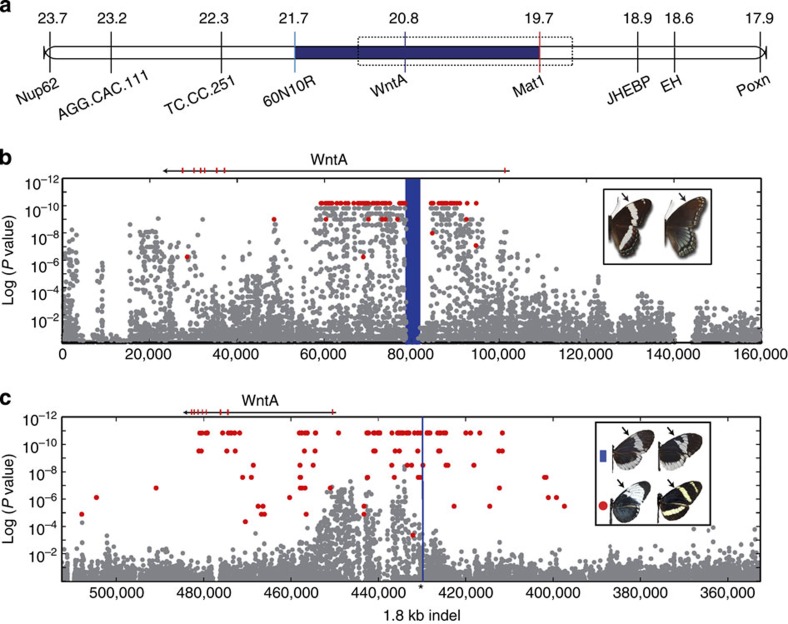
Figure 4. SNP associations with mimetic polymorphisms within Limenitis and Heliconius.
(a) The zero-recombinant colour-patterning interval in Limenitis (blue). (b) Variable SNPs (grey) mapped by position (x axis) against the P value for the Fisher’s exact test of allelic association with phenotype (y axis); SNPs fixed between mimetic and non-mimetic Limenitis individuals are shown in red. The highlighted blue interval corresponds to the position of a long interspersed element (LINE) retrotransposon fixed between mimetic and non-mimetic individuals. (c) SNPs mapped across the chromosome containing WntA in H. c. alithea (grey), with position on the x axis, and P value of Fisher’s exact test for allelic association on the y axis for the melanic/non-melanic phenotypes. There were no fixed SNPs between these two phenotypes; however, 170 fixed differences were detected among comparisons of allopatric H. c. galanthus and H. pachinus (red). Combined analysis of structural variation in H. c. alithea and all other Heliconius studies revealed a single 1.8-kb indel that was perfectly associated with variation in forewing melanin patterning (black arrows), which is positionally highlighted in blue for all species sequenced.