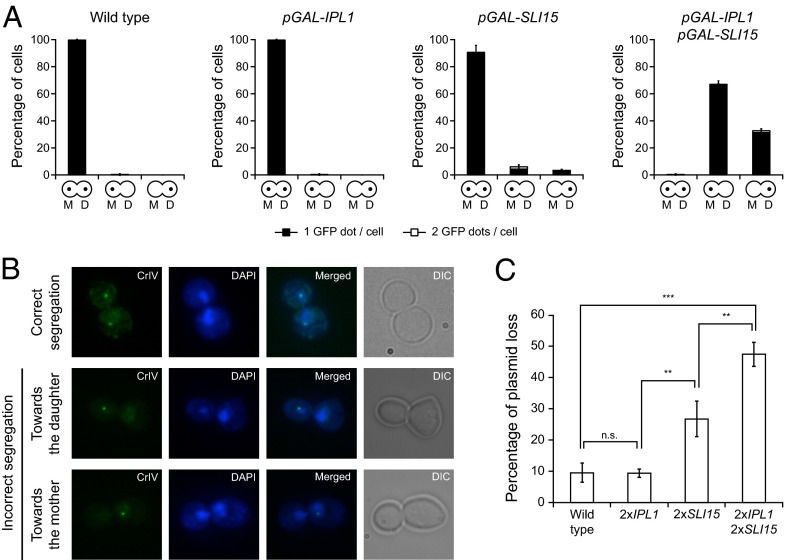
Fig. 3.
Ipl1 and Sli15 overexpression causes defects in chromosome segregation. (A and B) Wild-type (F955), pGAL-IPL1 (F256), pGAL-SLI15 (F953), and pGAL-IPL1 pGAL-SLI15 (F947) cells carrying a GFP tag on chromosome 4 (CrIV-GFP) were allowed to enter mitosis synchronously in YPRG as in Fig. 1A. (A) Analysis of chromosome segregation using CrIV-GFP dots. The graphics below the bars indicate whether sister chromatids segregated correctly [yeast cells with a dot in the mother (M) and a dot in the daughter (D) cell] or cosegregated toward the mother (a dot only in the mother cell) or toward the bud (a dot only in the daughter cell). Black bars indicate that only one GFP dot could be detected per cell; white bars indicate that two GFP dots could be detected per cell. Error bars indicate SD (n = 3). (B) Representative images showing the different patterns of CrIV-GFP segregation (green) and DAPI staining (blue). (C) Plasmid-loss assay with wild-type cells (F496) and cells carrying two copies of IPL1 (2×IPL1; F1753), two copies of SLI15 (2×SLI15; F1754), or two copies of both genes (2×IPL1 2×SLI15; F2019) under the control of their own promoters and also carrying the pRS316 plasmid. The graph shows the median values and SDs of three fluctuation tests for each strain; each test was performed using six independent colonies. Statistically significant (***P < 0.001 and **P < 0.01) or nonsignificant (n.s.) differences according to a two-tailed t test are shown.