Table 1. Mutations beneficial to ID8 under two selection regimes.
| Label | Selection |

|
Protein function | Protein name | Aa position |
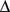 Aa Aa |
Nuc position |
 Nuc Nuc |
| T1 | 80°C | 1 | coat | F | 5 | T  A A |
2578 | A  G G |
| T2 | 80°C | 1 | coat | F | 355 | P  S S |
3628 | C  T T |
| T3 | 80°C | 1 | spike | G | 168 | R  C C |
4484 | C  T T |
| T4 | 80°C | 2 | spike | G | 38 | R  C C |
4094 | C  T T |
| P1 | pH 1.5 | 2 | coat | F | 77 | I  T T |
2792 | T  C C |
| P2 | pH 1.5 | 1 | spike | G | 65 | T  A A |
4175 | A  G G |
| P3 | pH 1.5 | 2 | pilot | H | 71 | A  V V |
4738 | C  T T |
The seven beneficial mutations identified in this study affected three different viral structural proteins: the major coat protein F, the spike protein G, and the pilot protein H. Positions are based on the published genome sequence of ID8 (GenBank accession # DQ079898). Nuc, nucleotide; n, number of lineages with mutation;  Nuc, nucleotide change; Aa, amino acid;
Nuc, nucleotide change; Aa, amino acid;  Aa, amino-acid change.
Aa, amino-acid change.
