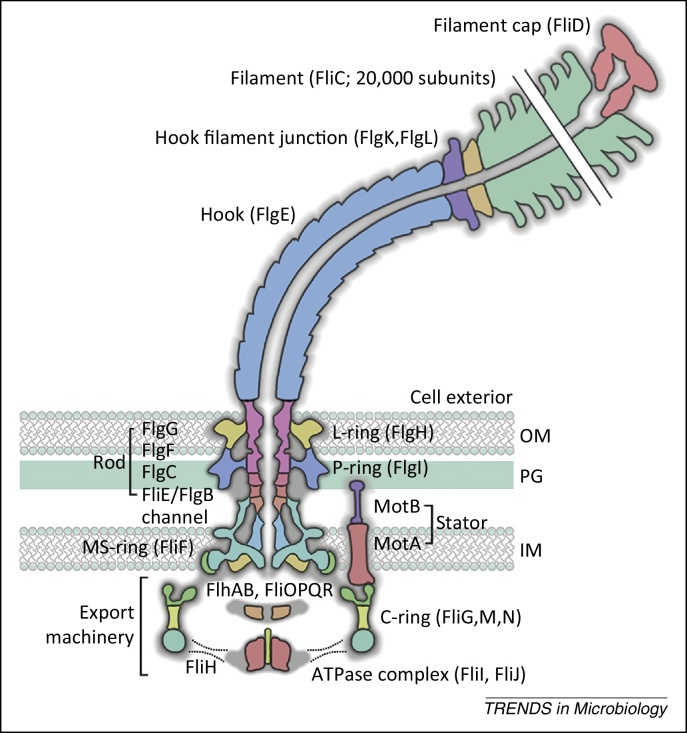
Figure 1.
The bacterial flagellum rotary nanomotor. The bacterial flagellum assembles from the inner membrane (IM) to span the PG cell wall and outer membrane (OM), finally extending into the extracellular space. Three contiguous hollow substructures – the rod, hook, and filament – are sequentially assembled. The drive-shaft rod (FliE, FlgB, FlgC, FlgF, and FlgG) is surrounded by a series of rings [lipopolysaccharide (L) ring, FlgH; PG (P) ring, FlgI; membrane–supramembrane (MS) ring, FliF; cytoplasmic (C) ring, FliGMN] and together these elements form the basal body. The C ring interacts with the stator units (MotAB) to drive flagellar rotation. The flexible hook (FlgE) extends from the cell surface with a defined length of ∼55 nm. Hook–filament junction proteins (FlgKL) connect the hook to the flagellar filament (flagellin, FliC). Subunits for the rod, hook, and filament are translocated across the cytoplasmic membrane by a dedicated type III export machinery that comprises an ATPase complex (FliI, FliJ, and FliH; the broken line is the predicted position of FliH), an unfolding cage (FlhAc), and a transmembrane export gate (FlhAB and FliOPQR). On crossing the membrane, subunits then transit through the central channel in the external flagellum to the distal tip, where they crystallise beneath specific cap foldases for the rod (FlgJ), hook (FlgD), and filament (FliD). The positions of individual protein structures are derived from cryoelectron tomography and represent the elements that remain in the mature structure.