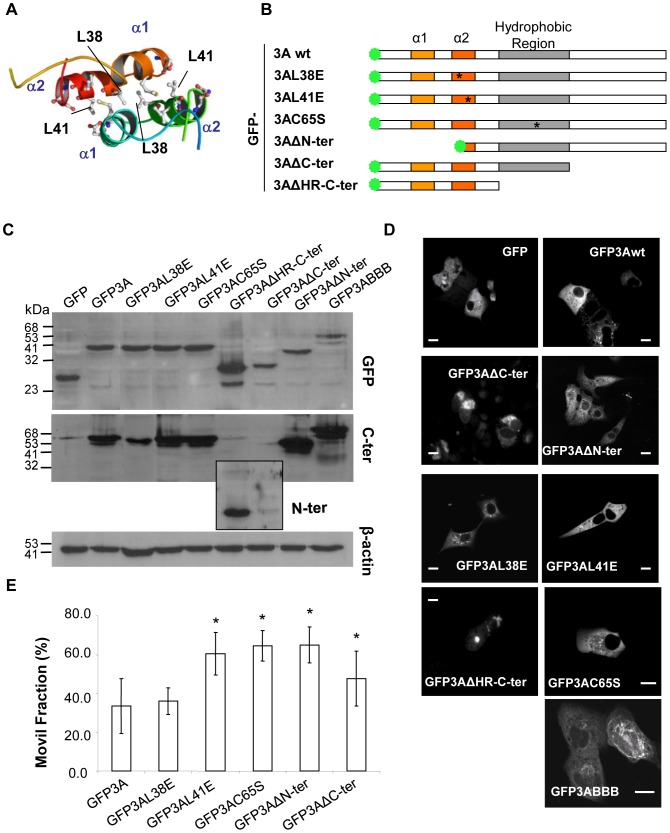
Figure 4. Analysis of GFP fusion proteins carrying mutations in 3A.
A) Structural model for FMDV 3A protein dimer (9). Ribbons represent α-helixes 1 and 2. Leucines at positions 38 and 41 are indicated. B). Schematic representation of the fusion proteins analyzed in which GFP (green stars), α-helixes (orange boxes) and the hydrophobic region (gray boxes) are indicated. Substitutions (L38E, L41E and C65S at residues conserved in 99, 99 and 85% among the FMDV sequences from the NCBI database, respectively) and deletions − ΔN-ter (I1-L41), ΔC-ter (R82-E153) and ΔHR-C-ter (K53-E153) −, generated as described in Materials and Methods, are shown. Asterisks denote single replacements. An alignment of the 3A sequences spanning the different mutations constructed among different FMDV serotypes can be found at [9] C) Western blotting of cells transiently expressing fusion proteins. Vero cells were transfected with 1 µg of plasmids expressing the fusion proteins indicated. Proteins were detected by incubation with a primary polyclonal antibody to the C-terminus (346) − with the exception of GFPΔHR-C-ter and GFPΔC-ter (shown boxed) that were blotted with serum 443 to the N-terminus − or with a MoAb to GFP. Blotting to an anti-β-actin was used as control of protein loading. Molecular weights are indicated in kDa. D) Fluorescent pattern of different GFP fusion proteins. Vero cells 24 h pt with the plasmids expressing the fusion proteins indicated were fixed and processed for confocal microscopy as described in Materials and Methods. E) Comparison of mobile fractions in FRAP of GFP3A fusion proteins. Vero cells were transfected as in (D), and 24 h pt FRAP was determined as described in the legend of Fig. 5. Data are presented as means ± the standard deviations of triplicate experiments (n>10). Statistically significant differences relative to GFP3A percentage of mobile fraction are indicated by an asterisk (P≤0.001). Scale bar, 20 µm.