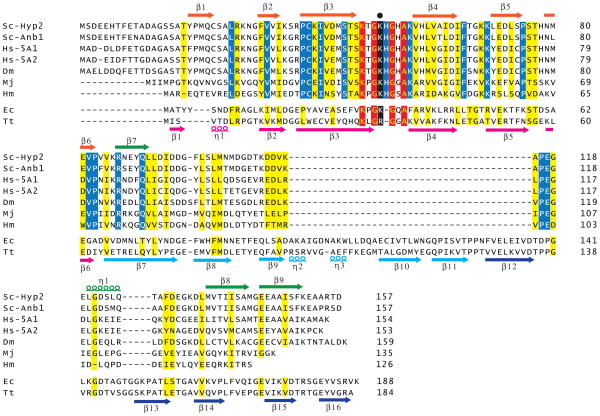
Figure 2. Sequence alignment of eIF5A and EF-P.
Multiple sequence alignment of yeast (Saccharomyces cerevisiae, Sc) Hyp2 (Tif51A; eIF5A1) and Anb1 (Tif51B, eIF5A2), human (Hs) eIF5A1 and eIF5A2, fly (Drosophila melanogaster, Dm) eIF5A, archaeal (Methanococcus jannaschii, Mj, and Haloarcula marismortui, Hm) aIF5A, and bacterial (Escherichia coli, Ec, and Thermus thermophilus, Tt) EF-P was performed using Clustal X (v2.0). Residues in red are identical in all eIF5A and EF-P proteins; residues in blue are identical only in eIF5A and aIF5A; and residues in yellow are conserved in eIF5A and EF-P. Secondary structure elements are shown at the top (eIF5A) and bottom (EF-P) of the sequences, and colored based on the protein domain and scheme described in Figure 3. The lysine residue that is modified to hypusine in eIF5A and aIF5A and β-lysinylated in E. coli EF-P is colored black and denoted by the black dot above the alignment. A color version of the figure is available online.