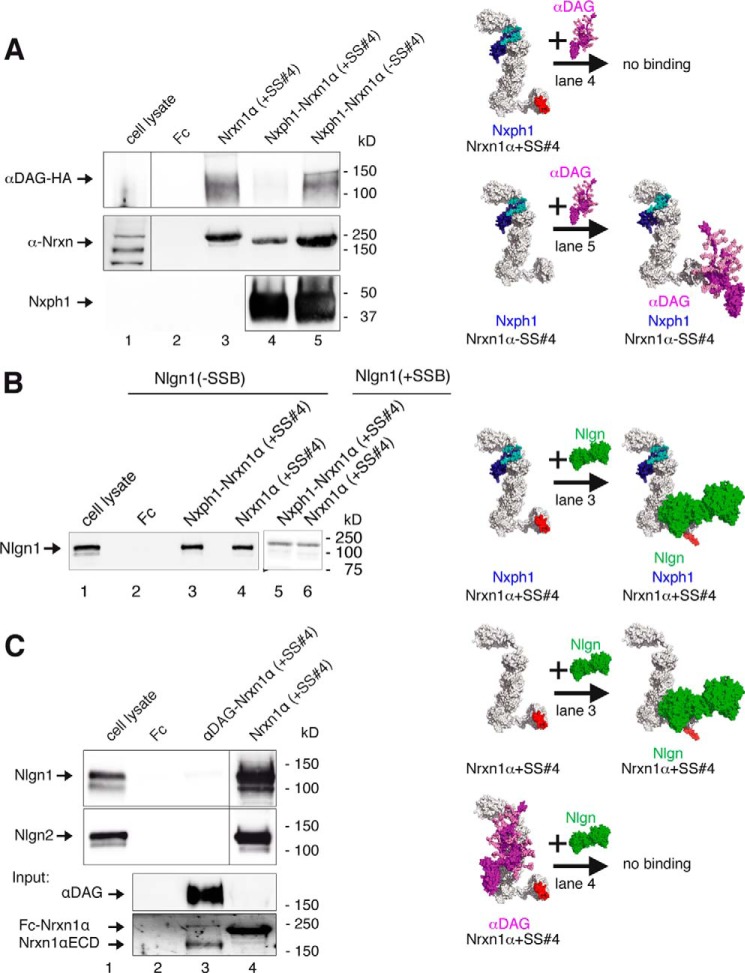
FIGURE 6.
Restrictions in α-Nrxn multiplexes with αDAG, Nxph1, and Nlgn. A, Nrxn1α triple complex with Nxph1 and αDAG depends on SS#4. Preformed complexes of Fc-tagged Nxph1 and Nrxn1α purified from HEK293 cell media were probed for binding of αDAG-HA from COS-7 lysate with (immunoblot, lane 4) and without (lane 5) insert at SS#4. A triple complex of αDAG·Nxph1·Nrxn1α only forms with LNS6(−SS#4) (see also Fig. 2B and Ref. 37). B, triple complex formation of Nlgn1·Nxph1·Nrxn1α has no splicing restrictions. Preformed complex of Fc-tagged Nxph1 and Nrxn1α binds to recombinant Nlgn1 (lane 3) even with inserts in Nrxn1α (+SS#4) and in Nlgn1(+SSB) (lane 5), similar to Nrxn1α alone (lanes 4 and 6). Binding of Nrxn1α+SS#4 to Nlgn1+SSB requires long incubation and exposure times (20, 25). C, αDAG and Nlgns do not bind simultaneously to Nrxn1α. Recombinant Nlgn1(−SSB) and Nlgn2 (upper and middle panels) bind to Fc-tagged Nrxn1α(+SS#4) (lane 4), but a preformed complex of αDAG with Nrxn1α (lane 3, lower panels) inhibits binding of Nlgn1(−SSB) or Nlgn2 (lane 3). All recombinant αDAG variants were purified from HEK293 cells co-transfected with LARGE; pictograms (A-C, right) visualize the complexes tested and key results (color-coded as labeled, splice inserts are in red). For αDAG representation, the model of De Rosa et al. (63) was extended with an elongated mucin region, and some recently determined O-linked glycans (100) were added. The size of molecules (right) are to scale, and the modeled complexes were generated using two criteria, (i) coverage of hot spots and (ii) maximal surface area buried (see “Experimental Procedures”). Note that in addition to complexes shown, other conformations are not excluded. The model of Nlgn/Nrxn1α was modified from Refs. 33 and 66.