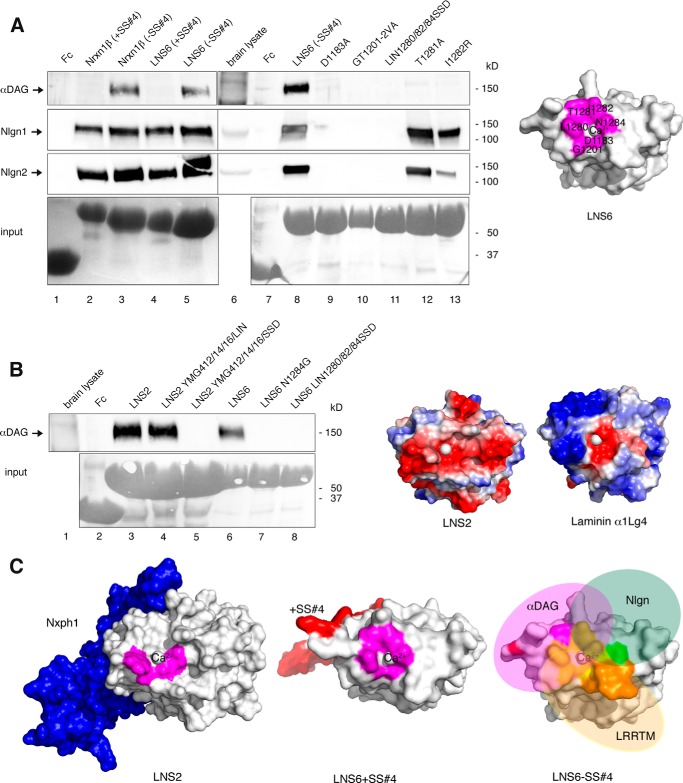
FIGURE 7.
αDAG competes with Nlgn for binding at Nrxn LNS6 domain. A, site-directed mutagenesis probing the binding epitope of αDAG at LNS6. The presence of splice insert +SS#4 in Fc-tagged Nrxn1β (lane 2) or isolated LNS6 (lane 4) and two point mutations in LNS6 (T1281A, lane 12; I1282R, lane 13) selectively block interaction with αDAG from brain lysate (upper panel) with unchanged Nlgn1 (second panel) and Nlgn2 (third panel) binding. Other residues at the calcium coordination site block binding to both αDAG and Nlgn (lanes 9–11), resulting in a partially overlapping epitope on LNS6 (magenta, right panel). B, αDAG binding requires calcium-coordinating and hydrophobic residues. Brain αDAG binds to normal LNS2 (lane 3) and a hybrid LNS with a triple mutation transferring the hydrophobic calcium coordination of LNS6 onto LNS2 (lane 4). Calcium coordination sites transferred from LNS2 to LNS6 (lane 7) or from laminin α2LG5 to LNS2 (lane 5) or to LNS6 (lane 8) abolish αDAG binding. The rim surface of Nrxn LNS2 domain is negatively charged (red, right panel), in contrast to laminin with more basic residues (blue, right panel). C, structural determinants of Nrxn-ligand interaction. Molecular modeling indicating that binding of Nxph1 at LNS2 (blue, left panel) and inclusion of splice insert at SS#4 in LNS6 (red, middle panel) may have a similar structural effect by sterically inhibiting the approximation of αDAG (right panel). In addition, binding epitopes for Nlgn (green), αDAG (magenta), and LRRTM (orange) overlap on LNS6 (yellow) but also have exclusive residues (right panel).