FIGURE 5.
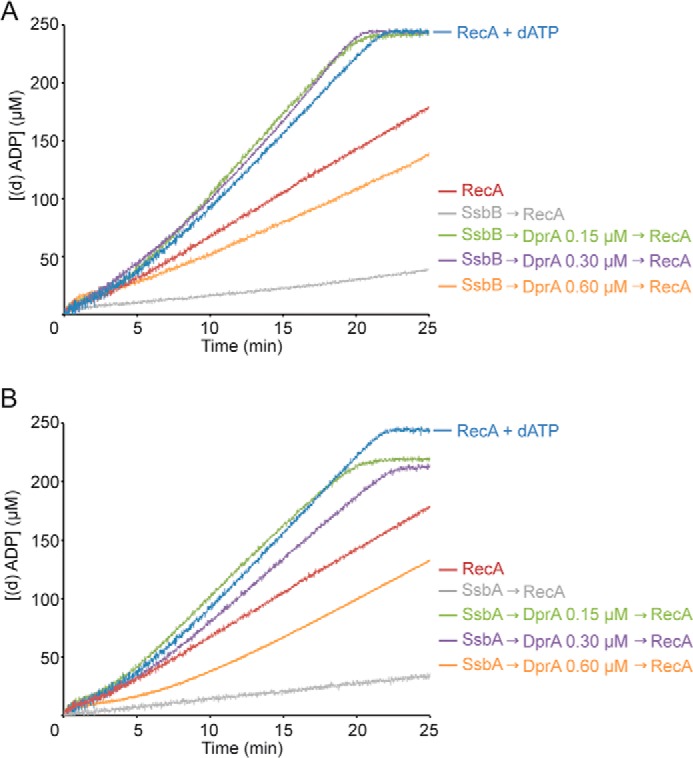
DprA overcomes the SsbA or SsbB inhibition for RecA-mediated ATP hydrolysis. A, circular ssDNA (10 μm in nt) was preincubated with SsbB (0.3 μm) for 5 min, and DprA (0.15, 0.3, or 0.6 μm) was added for 5 min at 37 °C in buffer A containing 5 mm ATP or dATP. Then RecA (0.8 μm) was added, and the ATPase activity was measured for 25 min. The activity of RecA in the presence of dATP was measured as control. B, circular ssDNA was preincubated with SsbA (0.3 μm) for 5 min, and DprA (0.15, 0.3, or 0.6 μm) was added for 5 min at 37 °C in buffer A containing 5 mm ATP or dATP. Then RecA (0.8 μm) was added, and the ATPase activity was measured for 25 min. The activity of RecA in the presence of dATP was measured as control. All reactions were repeated three or more times with similar results. The amount of nucleotide hydrolyzed was calculated as described in the legend to Fig. 3.
