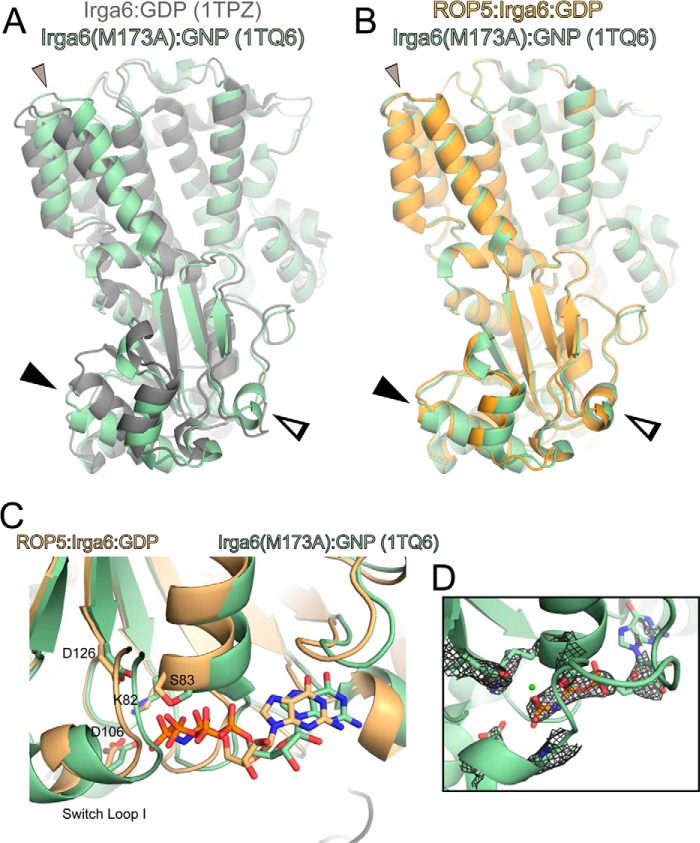
FIGURE 8.
IRGa6M173A occupies a conformation similar to that of ROP5-bound wild-type IRGa6. The previously published structure (Protein Data Bank code 1TQ6; green) of the mutant M173A Irga6 bound to GMPPNP (GNP) is overlaid with the published structure of Irga6-GDP (Protein Data Bank code 1TPZ; gray) (Cα RMSD of 2.5 Å over 393 atom pairs) (A) or ROP5-bound Irga6 (orange) (Cα RMSD of 1.9 Å over 383 atoms) (B). Closed arrowhead, helix 3; open arrowhead, switch loop I. Additional helical rotations are indicated with a small closed gray arrowhead. C, the IRGa6 active site of 1TQ6 is overlaid with that of the ROP5-bound structure. Residues that coordinate the nucleotide and associated waters are shown as sticks. D, the published structure factors for 1TQ6 were used to calculate the 2.7 Å 2Fo − Fc map, which is shown contoured at 1.5σ around the bound nucleotide. Note that although the 1TQ6 coordinates include a bound Mg2+, there is no density at the ion's position, nor is there evidence for the octahedral coordination typical of a Mg2+. This suggests that the conformation of M173A IRGa6 captured in this structure, like ROP5-bound IRGa6, excludes a highly ordered Mg2+ from the active site.