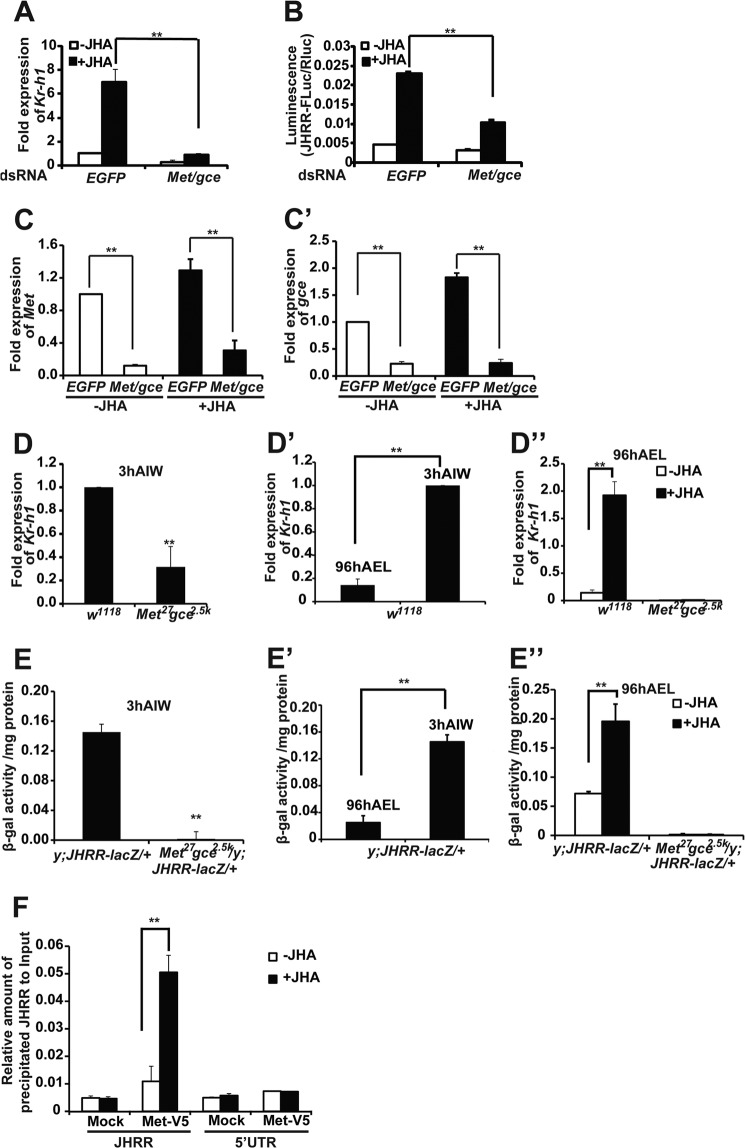
FIGURE 3.
Met and Gce are required for JH-induced Kr-h1 expression through JHRR. A and B, the JH-induced Kr-h1 expression and JHRR-driven luciferase activity were measured after Met and gce were simultaneously reduced by RNAi in Kc cells. EGFP RNAi was used as the control. A, for detecting the mRNA level of Kr-h1, Kc cells were treated with 1 μm methoprene (JHA) (DMSO as the control) for 30 min, and the relative Kr-h1 mRNA levels were measured by qPCR. B, for detecting the JHRR-driven luciferase activity, Kc cells were treated with 1 μm JHA (DMSO as the control) for 2 h, and then dual luciferase assays were performed. C and C′, RNAi efficiency of Met (C) and gce (C′) in Kc cells. D–D″) The mRNA levels of Kr-h1 in the fat body isolated from w1118 and Met27gce2.5k larvae at the indicated stage. 3 h AIW the JH titer is high. At 96 h AEL the JH titer is low. In D″, the isolated fat body tissues were treated with 1 μm methoprene (JHA) (DMSO as the control) for 30 min. E–E″, the JHRR-driven β-galactosidase (β-gal) activities were measured in the fat body isolated from the +/y; JHRR-LacZ and Met27gce2.5k/y; JHRR-LacZ/+ larvae at indicated stage. In E″, the isolated fat body tissues were treated with 1 μm methoprene (JHA) (DMSO as the control) for 2 h. F, ChIP. Kc cells were transfected with Met-V5 expression plasmid for 48 h followed methoprene (or DMSO as a control) treatment for 10 min. ChIP was performed using the PierceTM Agarose ChIP Kit (Thermo) and the V5 antibody to detect the binding between Met-V5 and the JHRR DNA or DNA in a 5′-UTR of Kr-h1α (in the 1st exon, lacking E-box-like motifs). The precipitated DNA and input were analyzed by qPCR. EGFP, enhanced green fluorescent protein.