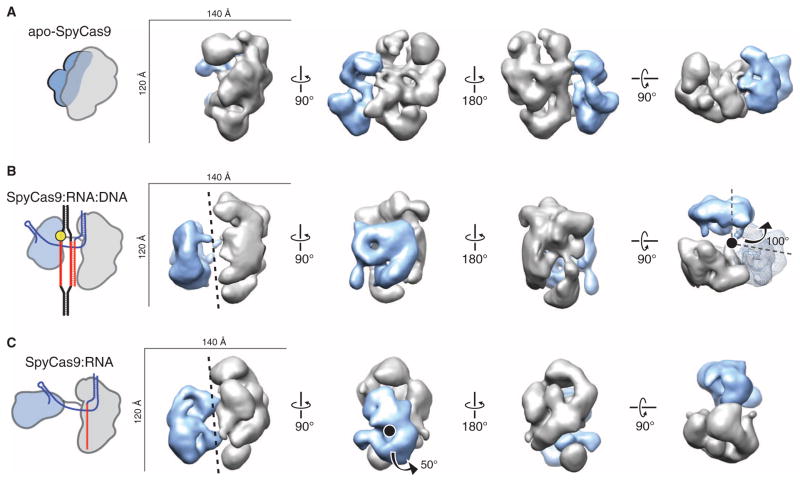
Fig. 5. RNA loading positions the two major lobes of SpyCas9 around a central channel.
(A to C) Single-particle EM reconstructions of negatively stained apo-SpyCas9 (A), SpyCas9:RNA:DNA (B), and SpyCas9:RNA (C) at 19-, 19-, and 21-Å resolution (using the 0.5 FSC criterion), respectively. Cartoon representations of the structures are shown (left). The structures are aligned on the basis of the optimal CCCs between the independent α-helical lobes (gray). The smaller RuvC lobe (blue) in SpyCas9:RNA:DNA and SpyCas9:RNA rotates by ~100° [arrow in (B)] with respect to this lobe in the apo-Cas9 structure (transparent mesh) to form a central channel (black dashed line). There is a ~50° rotation [arrow in (C)] of the smaller lobe of SpyCas9:RNA along an axis perpendicular to this channel relative to SpyCas9:RNA:DNA.