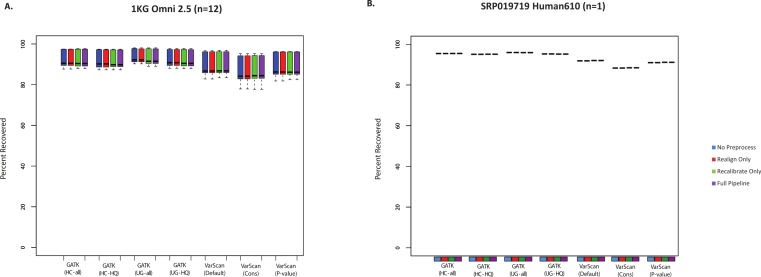
Figure 2.
(A) Recovery of variants reported from the Omni SNP chip for 1000 Genomes exome samples (n = 12). Only alleles that varied from the reference sequence and were located within the regions targeted in the exome sequencing design were considered for this analysis (this varied somewhat between samples, between a minimum of 477 variants and a maximum of 616 variants). Seven variant calling strategies were tested (GATK UnifiedGenotyper and HaplotypeCaller, with and without filtering low quality variants; VarScan with 3 sets of parameters, see Methods). “VarScan–Cons” is the most conservative set of parameters for VarScan. Each variant caller was also tested with 4 preprocessing conditions: variants called using both GATK indel realignment and quality score recalibration (“Full Pipeline”—purple), indel realignment only (“Realign Only”—red), quality score recalibration only (“Recalibrate Only”—green), or neither (“No Preprocess”—blue). (B) Same as A, but for 6437 variants on a different SNP chip design (Human 610) compared to exome variant calls for sample SRX265476.