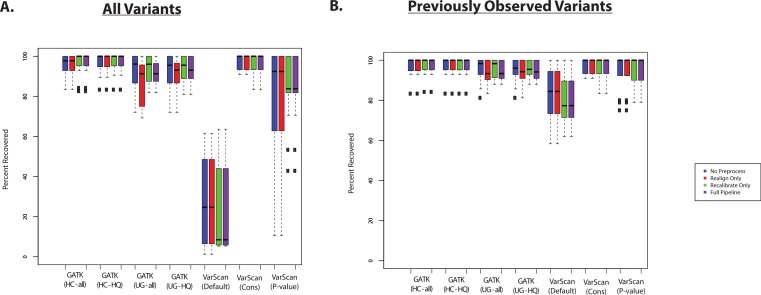
Figure 3. Recovery of coding SNPs from targeted exon samples (1KG Exome—chr20).
(A) SNP calls from paired targeted exon and exome datasets were compared to test the robustness of the calls made in the targeted exon data. Indel calls are not presented because there are almost no coding indels on chr20 for the targeted exon datasets. Two subjects have two targeted exon datasets (Tables S1 and S2), and concordance with exome datasets was reported for both targeted exon datasets separately (resulting in 14 concordance values per variant calling strategy). Seven variant calling strategies were tested (GATK UnifiedGenotyper and HaplotypeCaller, with and without filtering low quality variants; VarScan with 3 sets of parameters, see Methods). “VarScan-Cons” is the most conservative set of parameters for VarScan. Each variant caller was also tested with 4 preprocessing conditions: variants called using both GATK indel realignment and quality score recalibration (“Full Pipeline”—purple), indel realignment only (“Realign Only”—red), quality score recalibration only (“Recalibrate Only”—green), or neither (“No Preprocess”—blue). Concordance is reported as recovery of SNPs called in the targeted exon data, but these cannot be treated as “gold standard” variant calls. Most clearly, there was a high false positive rate when running VarScan with default parameters, so a high proportion of those variants called in the targeted exon samples could not be recovered in the exome dataset. In fact, on-target coverage is typically lower for the targeted exon samples than the exome staples (Tables S1 and S2). (B) Same as (A), but only previously observed variants are included in the percent recovery calculation.