Figure 6. Number of variants called for GATK versus VarScan (1KG—chr20).
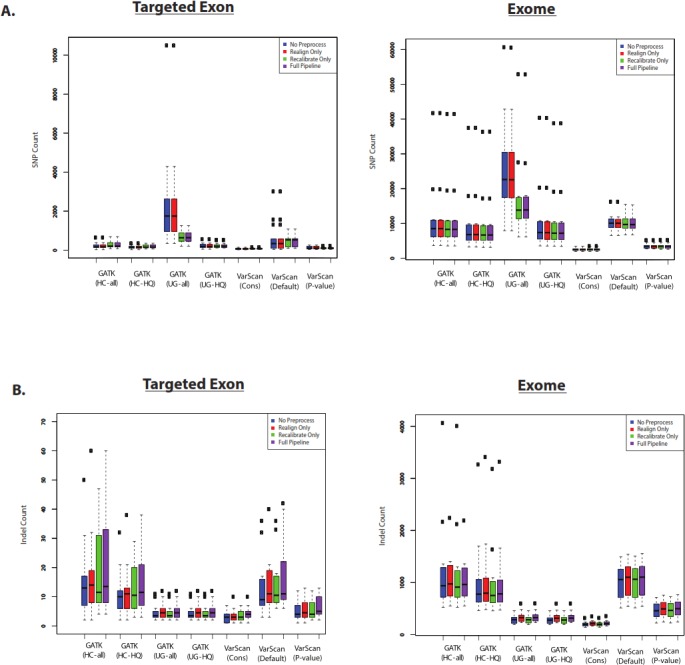
(A) Number of SNPs called for selected 1000 Genomes (1KG) targeted exon samples (n = 14) and exome samples (n = 12). The number of SNPs called is displayed for variants called using both GATK indel realignment and quality score recalibration (“Full Pipeline”—purple), indel realignment only (“Realign Only”—red), quality score recalibration only (“Recalibrate Only”—green), or neither (“No Preprocess”—blue). Variant counts are provided for the GATK UnifiedGenotyper (“UG”), GATK HaplotypeCaller (“HC”), and VarScan using 3 sets of parameters (see Methods). UnifiedGenotyper and HaplotypeCaller variants are then divided into the set of all variants (“UG-all” and “HC-all”) and higher-quality variant calls where variants flagged as low quality have been removed (“UG-HQ” and “HC-HQ”). “VarScan-Cons” is the most conservative set of parameters for VarScan. These values represent the total number of SNPs called. (B) Same as (A), for indels instead of SNPs.
