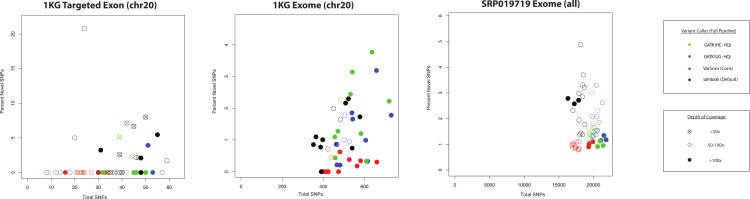
Figure 9. QC metrics to estimate specificity versus sensitivity for variant callers (coding SNPs).
For each of the three datasets characterized in this study (1KG targeted exon, n = 14; 1KG exome, n = 12; SRP019719 exome, n = 15), the number of coding SNPs called per sample is plotted along the x-axis and the proportion of novel variants is plotted on the y-axis. In order to simplify presentation of these results, we focused on the highest quality variant calls for each variant calling strategy: GATK UnifiedGenotyper with low-quality variants removed (UG-HQ, blue), GATK HaplotypeCaller with low-quality variants removed (HC-HQ, green), and VarScan using a custom set of conservative parameters (VarScan-Cons, red). Additionally, an unfiltered set of variants called via samtools are plotted in black. Only variants subject to GATK indel realignment and quality score recalibration (“Full Pipeline”) are considered for this comparison. The shape of the data point corresponds to the depth of on-target coverage: <50x coverage is represented as an X in an open-circle, 50–100x is represented as an open circle, and >100x is represented as a filled circle. If the novel percentage was tightly correlated with the actual false positive rate and the number of variants was tightly correlated with the actual sensitivity of the variant caller, than the ideal variant caller would show a cluster of data points in the bottom-right hand corner of the plot.