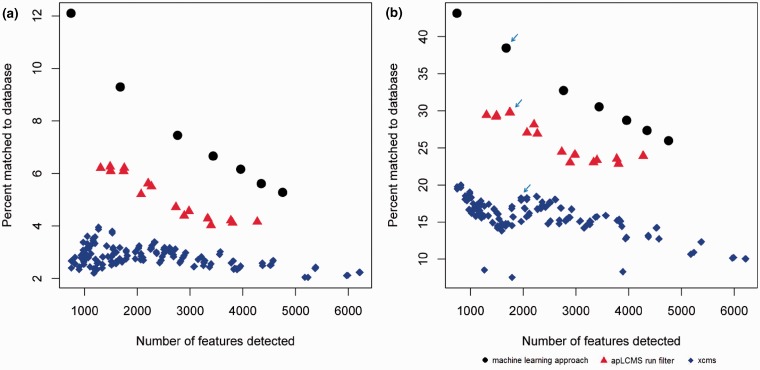
Fig. 3.
Comparing the percentage of peaks matched to known metabolite derivatives between the new machine learning approach against the existing run filter of apLCMS, and the matched filter of XCMS. All m/z values used in the training of the machine learning approach were removed. Orbitrap data generated from the NIST SRM 1950 samples was used. All three methods were allowed a number of parameter combinations. Each point represents a parameter combination. Matching was based on m/z value at the 5 ppm tolerance level. (a) Percent of newly detected features matched to the [M + H]+ ion forms of the half metabolites from HMDB held back from the methods. (b) Percent of newly detected peaks matched to [M + H]+, [M + K]+, [M + Na]+ or [M + NH4]+ ion forms in the MMCD. Arrows: data used in further analysis shown in Figure 4