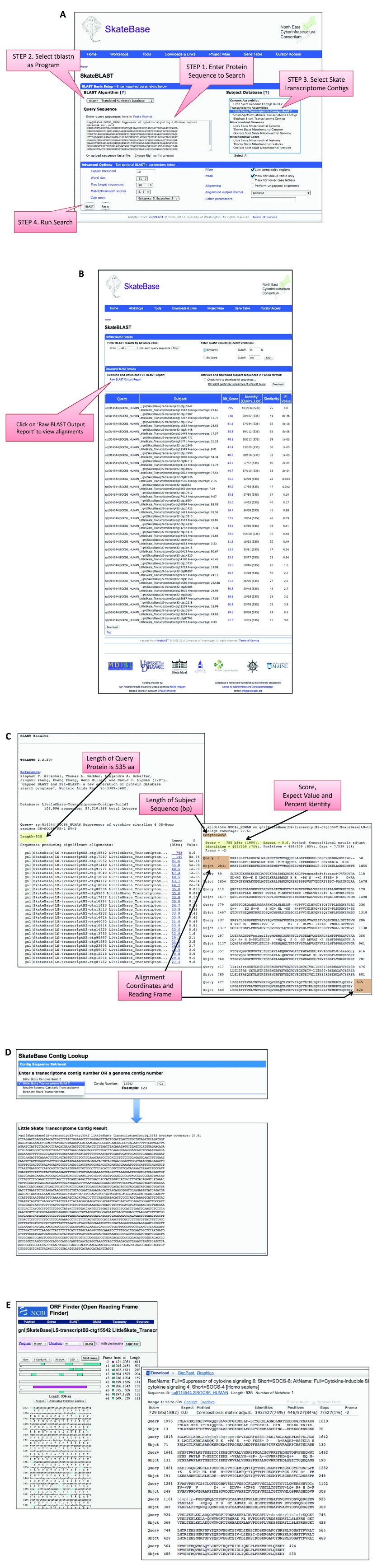
Figure 5. Example of using SkateBase and NCBI resources to find transcriptome data for SOCS6.
A. SkateBLAST query form showing the four steps to align the UniProt sequence for human SOCS6 (O14544) against the skate embryonic transcriptome using tblastn. Step 1 is to enter the sequence in FASTA format. The second step is to choose the tblastn program that will align the query protein sequence against translated sequences in all six possible reading frames. The third step is to select the embryonic transcriptome as the sequence database to search. The fourth step is to launch the search. B. The complete BLAST output can be accessed by clicking the “Inspect BLAST output” link at the top of the summary report page. This is necessary to examine the sequence alignments. C. Four important fields in the output should be examined carefully to interpret the alignments and determine which returned alignment best represents the skate ortholog to SOCS6. First, the alignment score, E-value, alignment length and percent identity can be used to interpret the overall alignment significance. Alignment coverage with respect to the query protein sequence and the subject transcriptome sequence can be interpreted by comparing the alignment coordinates to the length of the query protein sequence and length of the transcriptome sequence. In this example, the entire query protein sequence is covered by this transcriptome sequence. D. The SkateBase Contig Lookup tool can be used to retrieve the transcriptome sequence found in the SOCS6 tblastn search in FASTA format. Sequences from the skate genome assembly or the skate, S. canicula or C. milii transcriptome assemblies can be retrieved using this tool. E. Output from the NCBI ORF Finder tool showing a 536aa ORF in the skate transcriptome contig that best represents SOCS6 (left). Alignment from blastx search of the skate transcriptome sequence (contig 15542) against human UniProt using NCBI BLAST to validate that the contig aligned best to human SOSC6 rather than another human gene.