Table 2. Summary of Glycopeptide Tandem Mass Spectra for Intact-Mass Matched Compoundsa.
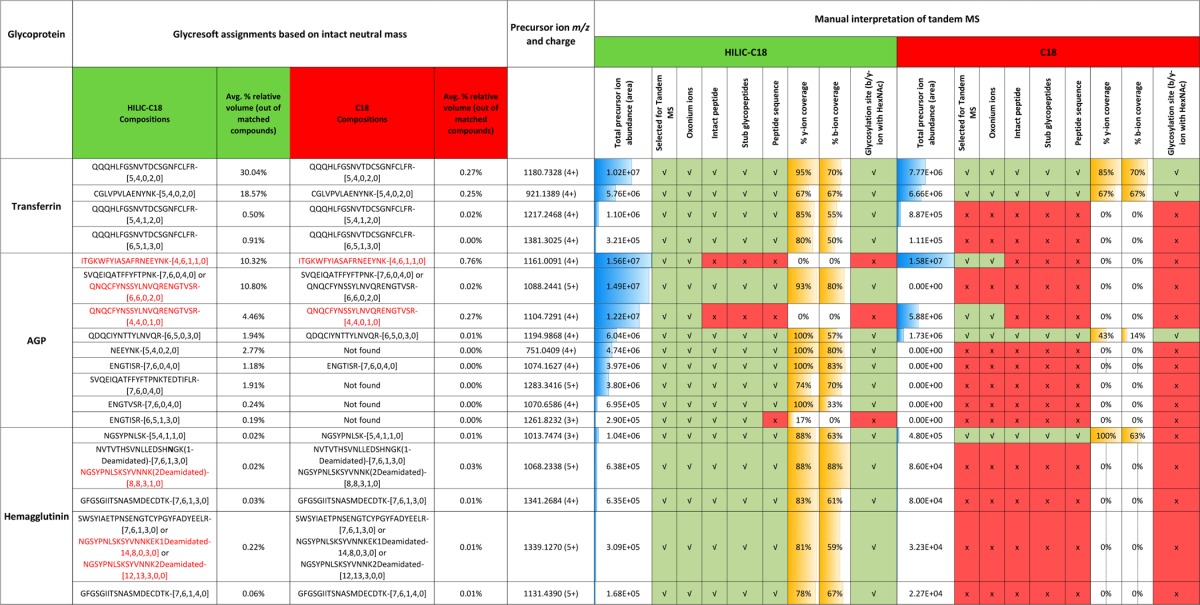
The features used in identification are listed in the table as yes/present (check mark) or no/absent (×). Abundances refer to the composite values of all charge states identified and matched for a given glycopeptide, presented as a percent of the total volume of all matched compounds. The absolute precursor ion abundances were determined from extracted ion chromatograms. Peptide backbone coverage was reported as percent product ions detected of total possible peptide product ions. Glycopeptide compositions in red indicate false/incorrect match. Glycopeptide nomenclature has been described in the text, and the peptide sequences are listed in Supporting Information Table S-1. Detailed mass tables confirming glycopeptide assignments have been presented in Supporting Information Section S-4. Glycopeptide identifiers are listed as peptide-[a,b,c,d,e], where a = number of hexoses; b = number of N-acetylhexosamines; c = number of deoxyhexoses; d = number of N-acetylneuraminic acids; e = number of N-glycolylneuraminic acids.
