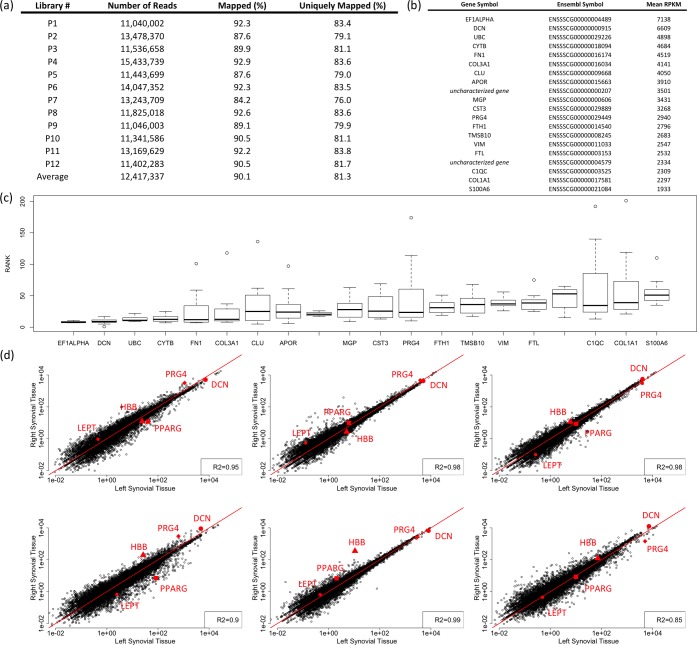
Figure 2.
RNA-seq of pig synovium is repeatable and indicates high expression of several transcripts encoding secreted proteins. (a) Table indicating the total number of reads and percentages of successfully mapped reads. More than 90% of reads on average are mapped to the pig genome (Susscr3), and more than 80% of reads are mapped uniquely. (b) Top 20 protein coding genes (based on expression level measured with RPKM) that do not originate from the mitochondria or the ribosome. (c) Box plot indicating the variation in the expression-based rank order of genes listed in (b) in all 12 libraries analyzed. The highest variation is in the rank of PRG4; however, it remains within the top 1% of genes in all libraries. (d) Scatterplots indicating high similarity (R2 > 0.85) between the synovial transcriptomes of paired left and right legs of all pigs. Data indicate RPKM, and each dot represents a single gene. Transcripts for the secreted proteins decorin (DCN) and lubricin (PRG4), red blood cell-derived beta hemoglobin (HBB), an adipocyte-derived transcription factor (PPARG), and secreted protein leptin (LEPT) are indicated with red colored symbols and closely follow the y = x line (solid red line) in each plot. Importantly, transcripts for matrix-degrading enzymes and inflammatory cytokines are not abundant, clustering in the bottom left corner of the panels.