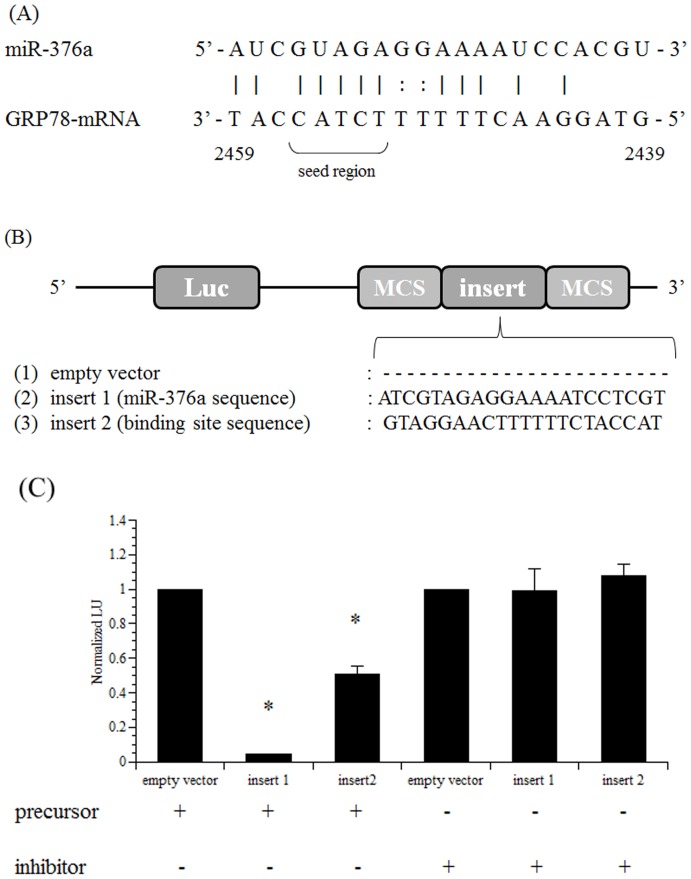
Figure 6. Luciferase assays for the identification of the rno-miR-376a-binding site in the 3′-UTR of GRP78 mRNA.
(A) Arrangement of rno-miR-376a and GRP78 mRNA and a schematic drawing of the predicted rno-miR-376a-binding site in the 3′-UTR of GRP78 mRNA. (B) Schematic drawings of the pMIR-REPORT luciferase vectors used in our experiment. To identify the rno-miR-376a-binding site in the 3′-UTR of GRP78 mRNA, luciferase reporter vectors were generated as described in the Materials and Methods. (C) Luciferase activity was measured to identify the rno-miR-376a-binding site in the 3′-UTR of GRP78 mRNA. HEK293 cells were prepared, and the cells were transfected with 200 ng of each reporter vector with 50 nM Pre-miR-376a (precursor) or Anti-miR-376a (inhibitor) as described in the Materials and Methods. For transfection normalization, the cells were also transfected with the pMIR-REPORT βgal vector. Luciferase activity was measured 24 h after transfection. The activity of the control (empty vector) was set at 1. Each value represents the mean ±SE of three independent experiments. *, significantly different from the control value, P<0.05.