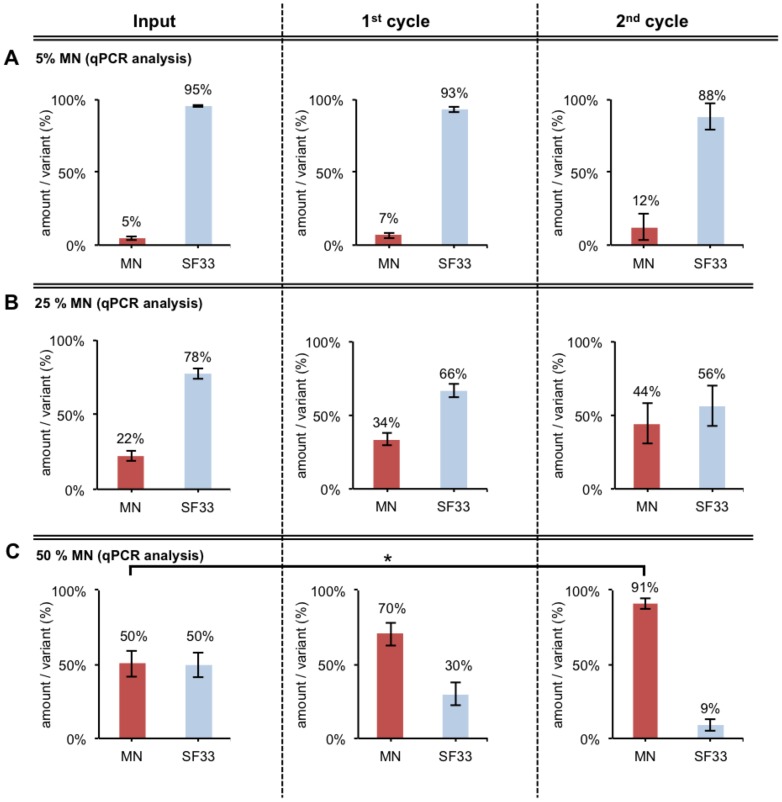
Figure 6. FACS-panning using different ratios of Env/V3-variants MN and SF33.
Different pQL9 Env/V3-variant plasmid mixtures containing A 5% MN/95% SF33, B 25% MN/75% SF33, C 50% MN/50% SF33 were prepared. HEK293T cells (13×106) were transfected (pQL9-mixture A, B or C respectively, pTN pack, pVSV-G) to form an un-linked virus library. Fresh HEK293T cells (13×106) were transduced at low MOI (0.1). The infected Env-genotype-phenotype “linked” cells were applied to the above-described FACS-sorting procedure 48 h after infection. The envelope genes were amplified from the genomic DNA of the collected cells, cloned into pQL9 and analyzed by qPCR. Fresh cells were transfected with the new pQL9 Env/V3-mixture (together with pTN pack and pVSV-G) to produce new virus for low MOI transduction of fresh cells and to enter another cycle of selection. The mean values of two independent experiments are shown. Statistics were calculated using the 1way-ANOVA followed by "Tukey's Multiple Comparison” test (* P<0.05; ** P<0.01; *** P<0.001). The relative amounts per variant of the input mixture, 1st and 2nd cycle are shown as analyzed by qPCR (Efficiency∧-Ct as % of each variant; triplicates) [69]–[71]. A reference primer pair that amplifies a sequence independently of the Env gene inserted was used to determine the total amount of pQL9 Env plasmids in each qPCR, respectively (data not shown).