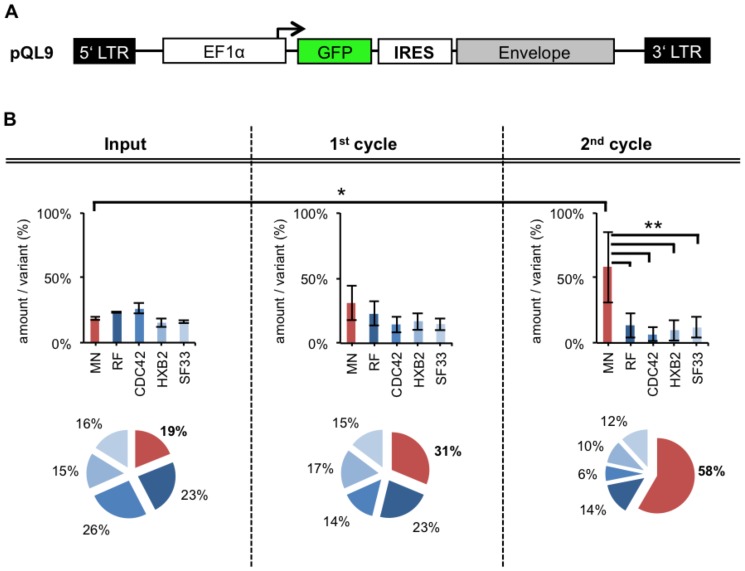
Figure 7. FACS-panning with pQL9 based Env/V3- model library.
A Schematic overview of pQL9 plasmid B HEK293T cells were transduced at low MOI (0,1) of “linked” pQL9 derived chimeric Env/V3-model-library viruses and subjected to a FACS panning and gating procedure 48 h after transduction (as described in Material and Methods and Figure 1/5). Env genes were amplified from genomic DNA of collected cells, cloned into fresh pQL9 and were analyzed by qPCR. New virus batches were produced with the resulting new pQL9 Env-library mixtures for infections of fresh cells, thus entering another cycle of selection. The mean values of four independent experiments are shown. Statistics were calculated using the 1-way-ANOVA- (testing, whether mean values differ) and “Dunnett's” post-test (testing, which mean values differ: * P<0.05; ** P<0.01; *** P<0.001). The relative amounts per variant of the input mixture, 1st and 2nd cycle are shown as analyzed by qPCR (Efficiency∧-Ct as % of each variant). The total amount of pQL9 Env/V3-plasmids in every sample was tested by a reference primer pair that amplifies a sequence independent of the Env genes, respectively (data not shown).