Figure 4.
Temporal Decomposition of Neutral and Deleterious Variation among Present-Day Individuals in Bottleneck Models
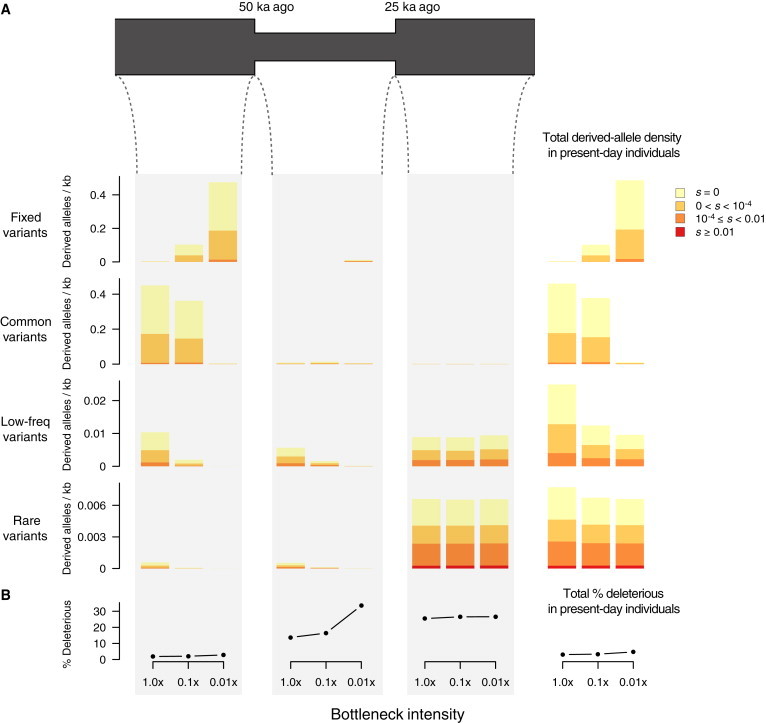
(A) Diagram of the simulated bottleneck model, in which population size decreased to 0.1× or 0.01× 50 ka ago and then recovered to prebottleneck levels with a size of 10,000 25 ka ago. Bar plots on the far right show the average number of derived alleles per individual per kilobase in present-day individuals as a function of DAF (fixed DAF = 1, common DAF ≥ 0.05, low-frequency 0.01 ≤ DAF < 0.05, and rare DAF < 0.01) and selection coefficients. We decomposed variant density in present-day individuals according to when the mutation arose (before, during, or after the bottleneck), as shown in the bar plots under the demographic model. Thus, we can obtain variant density of present-day individuals for each DAF category (rows) by simply adding variant density across the three time epochs considered.
(B) The proportion of derived deleterious alleles (|s| ≥ 10−4) per individual in present-day individuals and as a function of when the mutation arose.
