Abstract
Allorecognition has been well-studied in the context of vertebrate adaptive immunity and recognition of the Major Histocompatibility Complex (MHC), which is the central event of vertebrate immune responses. Although allorecognition systems have been identified throughout the metazoa, recent results have shown that there is no apparent conservation or orthologous relationship between the mechanisms underlying this phenomenon in different organisms. Thus the origin of the vertebrate adaptive immune system as well as these other complex recognition systems is a complete mystery. This review will focus on allorecognition in Botryllus schlosseri, a basal chordate which undergoes a natural transplantation reaction following contact between two individuals, and, analogous to vertebrates, is controlled by a single locus. We will summarize each of the known candidate genes within this locus and their potential roles in allorecognition, and speculate on how these findings may in fact be revealing potential functional relationships between disparate allorecognition systems.
Keywords: Allorecognition, Botryllus schlosseri, Invertebrate, fuhc, evolution, innate immunity
1. Introduction
Allorecognition, the ability to distinguish self from non-self cells and tissues between members of the same species, has been observed across the tree of life. However, only the Major Histocompatibility Complex (MHC) – based adaptive immune system has been well-characterized in jawed vertebrates. In organisms that predated the jawed vertebrates, nothing resembling the MHC has been discovered based on sequence homology. This raises the questions: where did the MHC originate and how did adaptive immunity evolve? Although the origin and evolution of the MHC remain an enigma, it has been postulated that the origins of MHC-based allorecognition may be discovered in primitive organism (Burnet, 1971). Botryllus schlosseri is a colonial ascidian (sea squirt), and a member of the subphylum Tunicata, the most primitive chordates. B. schlosseri undergoes a natural transplantation reaction which occurs at the tips of an extracorporeal vasculature (Bancroft, 1903). The outcome of allorecognition is dependent upon on a single highly polymorphic locus (Scofield et al., 1982a, 1982b) called the fusion/histocompatibility (fuhc) (Weissman et al., 1990). Individuals sharing one or both alleles at fuhc are compatible, while those that share neither are incompatible; a phenomenon that is reminiscent of the ‘missing self’ mechanism present in Natural Killer (NK) cells. Together, these characteristics make B. schlosseri an excellent model for investigating the mechanisms and evolution of allorecognition. In this review, we will focus on B. schlosseri allorecognition and the candidate players shown to be involved thus far, and attempt to put this system in a broader context of the evolution of allorecognition.
1.1 Botryllus schlosseri Life History
Tunicates occupy a key evolutionary position since they are considered the closest living relatives of the vertebrates (Delsuc et al., 2006). Embryogenesis results in a tadpole with a chordate body plan possessing a notochord, hollow nerve tube and a pharynx with gill slits. Shortly after hatching, the tadpole adheres to a surface, metamorphoses, and commits to a sessile invertebrate body plan. The founding individual (called an oozooid) will begin a life-long asexual budding process that will form a colony composed of numerous individuals (zooids) that are genetically identical. Each zooid has a complex body plan, including a heart, gastrointestinal tract, and nervous system. After several asexual budding cycles, the colony can become a fertile hermaphrodite. All of the zooids in a colony are interconnected with one another through a common extracorporeal vasculature which is encapsulated in a gelatinous, cellulose-based material called the tunic. At the periphery of the colony, the extra-corporeal vascular system forms finger-like structures called the ampullae. The tips of ampullae are composed of a single layer of columnar epithelial cells (Scofield et al., 1982b) and are the site of allorecognition.
1.2.1 Histocompatibility Outcome
When two colonies are in close proximity of each other, the ampullae may come into contact, and this will result in two possible outcomes: the ampullae will either undergo anastomosis, forming a vascular chimera (fusion), or undergo an inflammatory blood-based response resulting in the destruction of interacting ampullae, blocking further vascular interactions (rejection). The ampullae are large structures, outside the body and easily visualized and manipulated, and fusion and rejection is rapid, usually occurring within 24-48 hrs. - making this an excellent model system to study allorecognition responses. In an initial study by Bancroft (1903) on Botryllus and a related species in the genus Botrylloides, observations indicated there was ‘some similarity’ that allowed two colonies to fuse or reject, but believed age, size, and color were also contributing factors. Oka and Watanabe (1957) used Botryllus primigenus to follow up on Bancroft’s report and inferred that there was a single genetic factor determining histocompatibility. Using both wild-type and controlled crosses, the rules of fusion for B. schlosseri were found to be dependent upon a single highly polymorphic locus, which was named the fuhc (Sabbadin, 1962; Scofield et al., 1982a, 1982b; Weissman et al., 1990). Two individuals will fuse if they share one or both allele(s) of the fuhc locus and reject if they share no alleles. This mechanism resembles the ‘missing-self’ recognition system seen in NK cells (reviewed in Vivier et al., 2011) in vertebrates, where a target cell is compatible (i.e., it will not be killed) only if it expresses a self-marker. Similarly in B. schlosseri allorecognition, the recognition of a self fuhc allele will allow fusion between interacting colonies. In both NK cells and B. schlosseri allorecognition, failure to recognize self results in a cytotoxic outcome that will destroy the interacting/target cells.
The extent of polymorphism of the fuhc locus from and across different populations around the world was assessed by the frequency of fusibility between individuals collected within and across these populations (Boyd et al, 1990; Grosberg and Quinn, 1986; Rinkevich et al., 1991, 1992, 1995, 2001; Rinkevich and Weissman, 1991; Sabbadin et al., 1992). Rinkevich et al. (1991) estimated 479 to 560 fuhc alleles across three locations along the Mediterranean coast around Israel, and Grosberg and Quinn (1986) predicted 50 alleles along a 100 meter transect in Woods Hole, MA. To date, there is no evidence that B. schlosseri has recombination and diversification machinery homologous to those in vertebrates (e.g. Rag or AID), and thus it would appear that allorecognition is due to interactions between germline-encoded gene(s). This raises two interrelated questions: First, how can an innate allorecognition system discriminate between hundreds of fuhc alleles; and second, why are so many alleles maintained in a population?
1.2.2 Driving force for polymorphism: Germline Parasitism?
Buss (1982) proposed that somatic compatibility systems, like the one governing the natural transplantation seen in B. schlosseri, have evolved to prevent parasitism of the soma and germ line that could potentially occur after mixing the cells of two individuals. This phenomenon is well-documented and amenable to study in B. schlosseri. Following fusion of compatible individuals, Sabbadin and Zaniolo (1979) observed mobile germ cells being transferred from one partner to the other, such that both individuals in a parabiotic pair were harboring the others germline. This remained true even if the parabiosis was severed, demonstrating the presence of long-term germline progenitors which could migrate and contribute to development in another individual. In addition to the vascular-based natural transplantation of these cells, in some cases it was found that only a single genotype contributed to germline development in both individuals. Thus these germline progenitors appeared to be competing for germline niches in the newly developing buds, and the germline progenitors of one individual could out-compete and replace the germline of the other, in a process called germ cell parasitism (Sabbadin and Zaniolo, 1979). Subsequent studies examined somatic and germline parasitism between replicate chimeras (Laird et al., 2005; Pancer et al., 1995b; Stoner et al., 1999; Stoner and Weissman, 1996) demonstrating that the parasitism was repeatable, and further that a hierarchy of winners and losers in germline parasitism could be identified in both lab-reared and wild-type individuals. Importantly, the parasitic phenotype is retained during cell isolation and microinjection experiments, demonstrating that it is a property of the stem cells themselves, and not other factors transferred via the vasculature (Laird et al., 2005). Therefore, the generation of new self ligands and the ability to discriminate between them are likely a response to potential germline parasitism following vascular fusion, as Buss (1982) had predicted.
Parabiosis can also result in transplant of somatic progenitors, which can be visualized using extensive color polymorphism of pigment cells between different compatible genotypes. However, the extent of somatic chimerism and parasitism is not known. While self-renewing precursors to pigment cells can be prospectively isolated (Laird et al., 2005) it is unknown whether these are true hematopoietic stem cells that give rise to all blood lineages, as progenitor/progeny relationships for blood and other tissues have not been characterized. Interestingly, while investigating somatic chimerism, Sabbadin and Astorri (1988) noted that stable chimeras were able to change fusibility within a certain time-frame. For instance, if a stable chimera composed of individuals with fuhc alleles AC and AD interacted with a BD individual via the AD region of the chimera, then a rejection reaction would sometimes occur. These results paralleled the work of Taneda and Watanabe (1982) in B. primigenus, and imply that humoral factors influence the allorecognition reaction after formation of a vascular chimera.
One finding that has particular relevance to the evolution of allorecognition is that a parabiosis made between an adult and a newly metamorphosed juvenile can result in rapid germline chimerism in the adult (Brown et al., 2009). This has important evolutionary and ecological consequences for allorecognition. Sessile marine communities are highly competitive for space, with little room for larvae to settle and grow. In addition, sexual maturation takes about 8 weeks following metamorphosis. Thus by fusing into a sexually mature adult, a juvenile individual can be contributing to the gene pool 4-5X faster than settling alone and growing, without the associated risk of not surviving to sexual maturity. In addition, it has been demonstrated that larvae tend to settle near histocompatible individuals, raising their chances of fusion (Grosberg and Quinn, 1986). Together, this suggests that allorecognition events occur much more frequently than previously appreciated, which would drive the evolution of polymorphism. In addition, these interactions should provide testable hypotheses for the amount of germline chimerism in the field, as well as population structure, both of which we are currently investigating.
1.2.3 Initiation of Fusion/Rejection Processes
The early stages of fusion and rejection are visually similar. As two colonies grow towards each other, blood cells, primarily morula cells and some macrophage-like cells, can been seen accumulating within the juxtaposed ampullae in closest proximity. This accumulation occurs prior to any physical contact of the vasculature, implying that a secreted factor is initiating the response (Cima et al., 2004, 2006; Rinkevich et al., 1998). As the tunics come into contact, some tunic cells also migrate towards the interacting ampullae. Next, the tunics begin to fuse, and as they do, the two opposing ampullar tips grow closer, and the migration of morula cells to the juxtaposed ampullae continues. Within 24 to 72 hours after ampullar contact, the two ampullae with either fuse or reject one another.
1.2.3.1 Fusion
If the two opposing colonies share at least one fuhc allele, the interacting ampullae will fuse. Fusion is due to a process of remodeling in which the ampullae parabiose, then change their morphology to produce elongated blood vessels. After the new vessel is formed between the two colonies, macrophage-like cells are commonly seen, indicating that they may have a role in the ampullae and vascular remodeling, similar to vertebrate angiogenesis (Rinkevich et al., 1998). In vitro experiments demonstrate that macrophage-like cells are recruited to where morula cells are activated (Menin et al., 2005). Most likely, the mechanisms and signaling involved in angiogenesis and vascular remodeling in B. schlosseri are being utilized during the fusion process. However, no study to date has investigated the molecular mechanisms underlying this controlled parabiosis and de-differentiation.
1.2.3.2 Rejection
Rejection in B. schlosseri is characterized by the destruction of the interacting ampullae and the formation of a melanized scar called a point-of-rejection (POR). The rejection process is initiated primarily by the morula blood cell type (Sabbadin et al., 1992; Taneda and Watanabe, 1982), which migrates through the epithelial layer of the ampullae into the tunic matrix. During rejection, the permeability of the ampullar epithelium increases which was demonstrated experimentally by the leakage of India ink in B. primigenus (Taneda and Watanabe, 1982). Once in the tunic, the morula cells degranulate and release the content of their stored vacuoles containing phenoloxidases and polyphenols (Ballarin et al., 1995). The polyphenols are converted by phenoloxidases into quinones and semi-quinones that will auto-catalyze their conversion into melanin producing a dark brown scar characteristic of PORs. In the process, radical oxygen species (ROS) are produced that will damage nearby tissue and result in cell death (Ballarin et al., 1998, 2002). However, the rejection reaction is not binary. There is a range of rejection responses, from slight bleeding of the ampullae to complete amputation of the ampullae, first classified by Scofield and Nagashima (1983) using oozooids then later described by another classification system (Oren et al., 2008). The former study also demonstrated that the severity of rejection mapped to the fuhc locus (discussed further below).
Studies in Botrylloides simodensis demonstrated that their rejection reaction can also be stimulated by humoral factors isolated from incompatible blood plasma (Saito and Watanabe, 1984). In B. schlosseri when morula cells are incubated in vitro with blood plasma (cell-free) from incompatible individuals, degranulation is observed, whereas there are no significant morphological or enzymatic (phenoloxidase activity) changes when morula cells are incubated with autologous blood plasma. These results provide further evidence for a role of humoral factors in the rejection response (Ballarin et al., 1995, 1998; Cima et al., 2004, 2006). An incompatible humoral factor could explain the observation that for some incompatible pairings, fusion of the tunic does not always occur (Boyd et al., 1990) whereas studies on B. schlosseri from Venice revealed tunic fusion occurs regardless of compatibility (Milanesi et al., 1978). Nonetheless, upon activation of the morula cells in a rejection response, either in vitro or in situ, these cells will express cytokines that can be detected by antibodies raised against IL-1α or TNF-α (Ballarin et al., 2001, 2005; Cima et al., 2004, 2006; Menin et al., 2005). Menin et al. (2005) demonstrated by quenching the IL-1α-like and TNF-α-like with the corresponding antibodies, recruitment of macrophage-like cells can be prevented in vitro. Is the incompatible humoral factor the same as the ligand for allorecognition? Or is there another process that utilizes morula cells? In some xenorecognition reactions with other botryllids, a rejection response would occur (Hirose et al., 2002; Rinkevich et al., 1998; Saito, 2003) suggesting that there is an activation signal for rejection that is somewhat conserved in botryllids (reviewed in De Tomaso, 2009; discussed below).
2 Candidate proteins
Several attempts have been made to identify any ancestral immune genes (Danska, et al., 1990; Pancer et al., 1996; Fagan and Weissman, 1996; Khalturin et al., 2003; Gasparini, et al., 2008; Voskoboynik et al., 2013a) or related genes involved in allorecognition (Pancer et al., 1993, 1995a, 1997; Levi et al., 1997; Ballarin et al., 1999; Rosner et al., 2007; Oren et al., 2007) to examine the extent of similarities to the vertebrate system based on sequence homology. Although none of the genes discovered thus far are directly related to the MHC gene, several genes have been identified in the fuhc locus (Figure 1) with potential roles in the allorecognition response. Among them is cfuhc genes (De Tomaso et al., 2005) which have been proposed to be the ligand(s) of a fester family (McKitrick et al., 2011; Nyholm et al., 2006) based receptor.
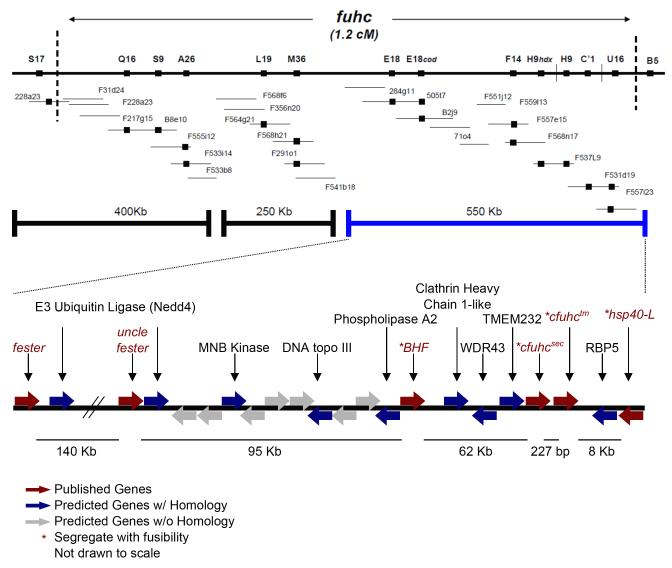
Figure 1.
Schematic of the fuhc locus. Top is a genetic map, with the genomic clones making up the minimal tiling path of the physical map shown below. Specific contigs are represented by bold lines, and the blue line shows the contig in which all polymorphic genes have thus far ben discovered. Predicted genes located in the locus are shown below. Red arrows are putative allorecognition genes, blue arrows are genes with homologs in the NCBI database (with a threshold of e-10), and gray arrows are predicted genes unique to Botryllus. The dotted vertical lines mark the boundaries and genetic markers within a 1.2 cM distance. Black solid boxes represent genetic markers.
2.1 cFuhc
2.1.1 The Search
Histocompatibility in B. schlosseri is clearly due to the segregation of a single locus, as fusion/rejection responses segregate in simple Mendelian ratios, even in wild-type crosses (Scofield et al., 1982a). Using strains of known specificity, the fuhc locus was isolated using forward genetics. Genetic mapping was done using several independent crosses, which delineated the region to a ca. 1 cM region of the genome. Genomic walking using these markers isolated 4 contigs and totaled 1.2 Mbp and covered both chromosomal breakpoints, although the size of the gaps between them is not known (Figure 1; De Tomaso et al., 1998; De Tomaso and Weissman, 2003a, 2003b; De Tomaso et al., 2005). Thus far, all genes with characteristics suggesting their role in histocompatibility have been found on a single 470 Kb contig (De Tomaso et al., 2005), which lies between contigs containing the chromosomal breakpoints. While the genome of B. schlosseri has recently been published (Voskoboynik et al., 2013a), the majority of the scaffolds are < 5 Kb, and none have been found that bridge the gaps in the physical map of the fuhc locus. Thus the locus has not been completely isolated and characterized yet, and it is quite possible that more genes involved in this process have not been discovered.
The first polymorphic gene identified which segregated with histocompatibility outcome was the cfuhc (De Tomaso, et al., 2005). While our original working hypothesis was that the cfuhc may be the ancestral MHC (Scofield, et al., 1982), it was found that the cfuhc is not related to the MHC by sequence homology or conserved protein domains. The cfuhc gene spanned 33 kb and consisted of 31 exons that were originally thought to be alternatively spliced into two mRNAs, the first encoding a secreted form (555 amino acids, encoded in exons 1-17) and a large transmembrane form (1008 amino acids, encoded in exons 1-14 spliced to 18-31). A later study (Rinkevich et al., 2012) indicated that the cloning and amplification of the transmembrane cfuhc could not be reproduced and claimed that polymorphisms did not correlate with the allorecognition response, concluding that another gene must be the allodeterminant. Indeed, initial assessment of cfuhc was incorrect and the cfuhc locus actually contains two genes: a 555 amino acid secreted form (cfuhcsec) and a type I 531 amino acid transmembrane form (cfuhctm) (Nydam et al., 2013c). The two genes are separated by a 227 bp intergenic region (Figure 1), but transcripts containing exons of both genes can be identified (Nydam, et al., 2013). However, there are no predicted promoter or splice leader sequences present for the transmembrane form. This could imply a new mechanism of transcriptional regulation via trans-splicing, which has been described in tunicates previously (reviewed in Hastings, 2005). However, it seems more likely that some of the transcripts identified in the initial cfuhc report (De Tomaso et al., 2005) were due to RNA polymerase reading through the short intergenic region, as single transcripts encoding exons from both genes were identified using independent analyses, including RT-PCR, RACE and mRNA-Seq (Nydam et al., 2013c), however, they are rare. Additionally, the two genes (cfuhcsec & cfuhctm) still correctly predict histocompatibility outcome in an independent study (Nydam et al., 2013c) and are the most polymorphic genes in the locus (Nydam et al., 2013a), but whether or not one is the allodeterminant or how each form biochemically contributes to the allorecognition reaction remains unclear.
2.1.2 Localization
The expression patterns of cfuhcsec and cfuhctm remain controversial due to discrepancies between studies (De Tomaso et al., 2005; Nydam et al., 2013c; Rinkevich et al., 2012). In De Tomaso et al. (2005) and Nydam et al. (2013c), expression of cfuhcsec and cfuhctm were in the epithelium of the ampullae along with a subset of blood cells. Conversely, in Rinkevich et al. (2012), the authors did not detect any expression in the ampullae, but did detect expression in a subset of blood cells. Regardless of the expression patterns established by in situ hybridization analysis, each study was able to detect parts of cfuhctm and cfuhcsec by RT-PCR or qPCR from ampullae tissue. Finally, the two genes are not expressed at equivalent levels. Based on the qPCR data and the expression pattern of the in situ hybridization presented in Nydam et al. (2013c), cfuhcsec appeared to be expressed at a significantly higher level compared to the transmembrane form in ampullae tissue.
2.1.3 Structure
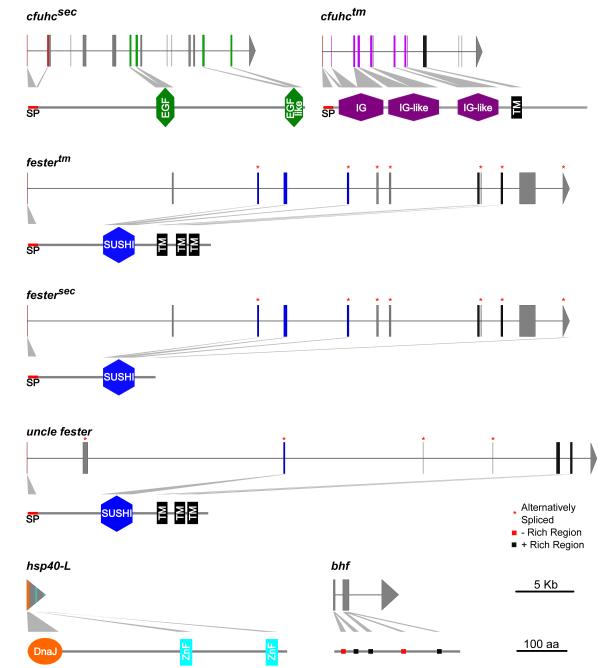
As shown in Figure 2, the cfuhcsec possesses a signal peptide based on analysis from SignalP 4.1 (Peterson et al., 2011) and two EGF-like domains (amino acid (a.a.) position 252-295 & 514-551) by ScanProsite (De Castro et al., 2006). Meanwhile cfuhctm has a signal peptide sequence until a.a. position 20 (Val) (Peterson et al., 2011) and is predicted to have two Ig-like domains spanning a.a positions 34-121 and 136-232 (De Castro et al., 2006) along with a transmembrane domain from a.a. 377 to 401. SMART (Letunic et al., 2012) predicted an EGF domain (a.a. 262-295) and an EGF-like domain (a.a. 517-551) in cfuhcsec and an Ig domain (a.a. 25 to 133), two Ig-like domains (a.a. 139-228 & 276-348) and a transmembrane domain (a.a. 377 to 399) in cfuhctm (Figure 2). Across these two genes, Nydam et al (2013a) found that exons 5 and 6 of cfuhcsec and exons 4 (part of the Ig/Ig-like domain) and 11 (transmembrane domain) of cfuhctm were under selection, but whether these exons are under balancing or directional selection remains unclear. Regions that are under selection are most likely structurally and functionally important to elucidate how this polymorphic protein is discriminated.
Figure 2.
Exon/intron organization and corresponding protein structures of the known allorecognition genes in B. schlosseri. SP = Signal Peptide, TM = Transmembrane, ZnF = Zinc Finger.
2.2 fester Family
Two of the genes present in the fuhc locus, fester (Nyholm et al., 2006) and uncle fester (McKitrick et al., 2011), are putative receptors in the allorecognition system. These two genes share over 90% identity on the nucleotide level for the last 585 bp with >70% identity on the amino acid level towards to C-terminal (McKitrick, 2011). They share homology between exons 6-11 for fester and exons 4-9 for uncle fester.
2.2.1 Fester
2.2.1.1. Genetics & Structure
Shortly after the putative ligand cfuhc was published, a putative receptor, fester, was reported (Nyholm et al., 2006). fester is located ca. 200 kb upstream of the cfuhc locus (Figure 1) and contains 12 exons spanning across 45 kb (Figure 2). The transcript can be alternatively spliced forming a secreted or transmembrane protein. Through RACE, a 2.3 kb cDNA was identified predicting a multi-pass transmembrane protein (Type IVa) with a signal sequence followed by a short consensus repeat (SCR) at the N-terminal end. The SCR domain is located in exon 5 while exons 8-10 each code a predicted transmembrane domain. The secreted form can be spliced together by attaching the two amino acid long exon 12 before the predicted transmembrane domains (Figure 2). The initial study on fester genetics (Nyholm et al., 2006), found that exons 3, 5, 6, 7, 9, 10, and 12 could be alternatively spliced in all combinations, putatively producing 36 splice isoforms. The theoretical maximum from these studies predict 80 different splice isoforms, where 64 isoforms are transmembrane while the remaining 16 are secreted variants. One of the most intriguing aspects of fester alternative splicing is that it appears to be individual specific. Cloning and sequencing of fester isoforms from any individual produces a unique repertoire of splice variants, from 8-24 per individual. Each individual also appears to express a full-length transmembrane form. This was done using non-quantitative methods which could have strong PCR and cloning bias, however initial analysis of mRNA-Seq data supports these results (Taketa et al., unpublished). Individual-specific alternative splicing suggests a role in specificity of Fester during allorecognition responses, and was the first indication that this may be a receptor.
This splice variation also has an interesting topological effect. Exons 8-10 each code for a predicted transmembrane domain, and can be alternatively spliced out. Several of the identified splice variants would actually change the topology of the protein, such that the intracellular end of the protein, encoded on exon 11, could be relocated to the extracellular side of the cell in certain isoforms. However, no direct evidence supports this topology prediction. If the intracellular domain of Fester is localized to the extracellular region, then the function of the intracellular region, if any existed, would be abolished. While there are no known signaling motifs encoded on this region of the protein, this is puzzling unless signaling is dependent upon the loops between each transmembrane domain interacting with an adaptor. Exon 8 encodes a true TM domain as determined by cell surface expression of a construct encoding exons 1-8 spliced to exon 11, thus it could also be that the predicted alpha helices encoded on exons 9 and 10 are not true transmembrane domains.
The fester gene is highly polymorphic, but not as polymorphic as the cfuhc gene (Nydam and De Tomaso, 2012; Nyholm et al., 2006). Most of the polymorphisms result in non-synonymous changes in the extracellular portion (Nyholm et al., 2006) and are clearly under balancing selection (Nydam and De Tomaso, 2012) which means there is more variation within population than across different populations. In addition, based on the amount of non-synonymous changes on the amino acid level and sampling along to coast of California, phylogenetic analysis of the extracellular region generated three clades (A-C) (Nyholm et al., 2006). Additionally, utilizing known fuhc crosses from De Tomaso et al. (2005), neither fester’s allele nor haplotype determined histocompatibility outcomes. For example, two individuals sharing the same fester haplotype can still reject one another.
2.2.1.2 Localization
Expression of fester was seen in the blood and ampullae by RT-PCR and RACE where different isoforms can be isolated. Through whole mount in situ hybridization, signal was detected in some blood cells and epithelium of the ampullae in adults, and the adhesive papillae and developing ampullae of the tadpole. The localization by in situ hybridizations was further verified by whole mount immunohistochemistry (IHC) using a monoclonal antibody (mAb) against Fester A allele. IHC displayed signal in the same areas seen by in situ hybridization. The blood cells positive for fester expression were believed to be macrophage-like cells and/or signet ring cells but morula cells, the effectors of rejection, did not appear to express the gene (Nyholm et al., 2006). Collectively, the data indicate that fester is localized to the site of allorecognition, the ampullae.
2.2.1.3 Function
The role of fester in allorecognition was assessed through two methods: siRNA knockdown and monoclonal antibody interference. Through siRNA knockdown of fester, histocompatible pairs resulted in a ‘no reaction’ phenotype. The same phenotype was seen if the knockdown was done between incompatible pairs. This implied that fester was involved in the initiation process. Further evidence of fester’s role in allorecognition was provided by antibody interference experiments. When the antibody against Fester clade A (FesterA/A) protein was injected into histocompatible pair (FesterA/A), then a fusion occurred as expected. In contrast, when a FesterA/A individual was injected and paired with an incompatible FesterA/B, rejection was overturned into a fusion (Nyholm et al., 2006). This suggested that binding of the mAb mimicked Fester binding to a ligand. This response was allele-specific, as mAb injection had no effect on rejecting colonies if neither expressed the Fester A allele. More importantly, if the antibody was injected into a Fester heterozygote (FesterA/B) paired with an incompatible Fester heterozygote (FesterA/B), then rejection was not affected. This demonstrated a dosage-dependent response to the mAb binding. Collectively, these two experiments indicated that fester may have multiple roles in the allorecognition process, one in initiating the rejection response, and the other in recognition of the cFuhc ligand.
Are the roles for Fester mediated by the repertoire of isoforms, the expression of the transmembrane and secreted, or the combination of the two? One possibility is that a complex of Fester isoforms, each with a different affinity for the histocompatibility ligand, has been assembled on the cells surface, such that the overall avidity is above a threshold for detecting self vs. non-self binding. This would be equivalent to stochastic expression of inhibitory receptors for MHC Class I molecules in NK cell education (reviewed in Vivier, et al., 2011). The secreted Fester isoforms could be humoral factors if they are localized to the vasculature. An alternative role for the secreted forms could be modulating the sensitivity or specificity of the transmembrane form if it they are localized along the membrane of the ampullar epithelia.
2.2.2 Uncle Fester
2.2.2.1 Genetics & Structure
The uncle fester locus is located ca. 140 kb downstream of the fester locus but ca. 150 kb upstream of the cfuhc (Figure 1) and appears to be a duplicate of fester exons 6-11 and surrounding regions. Through RACE, cDNA clones indicated that there are 9 exons spanning across 46 kb of genomic sequences transcribing a ca. 1.1 kb coding sequence translating into a 363 a.a. protein. Interestingly, unlike the other candidate allorecognition genes, uncle fester is not polymorphic and produces fewer splice isoforms compared to fester. From RT-PCR, only 9 isoforms of uncle fester were identified, with 3 variants expressed by the adult (full length, no exon 5, and no exons 4 and 5) and 6 variants expressed only by embryos and tadpoles (combination of exons 2, 3, or 4 and 5 are spliced out). The predicted protein contains a signal sequence, an SCR domain, and 3 transmembrane domains, classifying uncle fester as another Type IVa transmembrane protein. Exons 1-3 are unique to uncle fester (McKitrick et al., 2011), where exon 3 codes a SCR domain (a.a. 148-210) (De Castro et al., 2006) while exons 6-8 each code for a transmembrane domain (Figure 2).
2.2.2.2 Localization
uncle fester was initially characterized during embryogenesis by RT-PCR. In conjunction with whole mount in situ hybridization, expression of uncle fester was found in the ampullae and adhesive papillae of the tadpole as well as the ampullae and a population of blood cells in adults (McKitrick et al., 2011). Expression of uncle fester at those sites was confirmed by whole mount immunofluorescence using an antibody raised against the full-length protein. The antibody does not cross-react with Fester protein (McKitrick et al., 2011). Therefore, the antibody most likely has specificity for the unique region of Uncle Fester (exons 1-3). Using the monoclonal antibody from Nyholm et al. (2006) against Fester in conjunction with the antibody against Uncle Fester (McKitrick et al., 2011), populations of blood cell that expressed Fester, Uncle Fester, or both were identified using both immunofluorescence and by flow cytometry (McKitrick et al., 2011). However, due to the lack of hematopoietic lineage markers, the type of blood cells that differentially express fester or uncle fester or both is unknown, and the function of each population of cells is unknown.
2.2.2.3 Function
Similarly to fester, the function of uncle fester was determined by siRNA knockdown and by antibody inference. In the siRNA knockdown experiments, knockdown of uncle fester between two incompatible individuals resulted in a ‘no reaction’ phenotype. Meanwhile, if knockdown of uncle fester between two compatible individuals occurred, then a fusion will still occur (McKitrick et al., 2011). This indicated that uncle fester is solely involved in the rejection pathway and that fusion and rejection are not mutually exclusive. The antibody inference experiment demonstrated that upon contact of the Uncle Fester antibody to an ampulla, a rejection response will occur, even in the absence of another individual. Additionally, if the Uncle Fester antibody was placed between two colonies that were about to undergo a fusion reaction, the fusion would be prevented and a rejection response would be elicited instead (McKitrick et al., 2011). One hypothesis proposed in the study (McKitrick et al., 2011) was Fester and Uncle Fester heterodimerize to form the receptor for rejection. This may be true for the cells in the epithelium of the ampullae; however, the role of these proteins in the blood cells is still unknown.
2.3 Hsp40-L
Approximately 8 kb downstream of the 3’ end of the cfuhc transmembrane gene, the hsp40-l gene is coded in the opposition direction (Figure 1). This 2095 bp intron-less gene codes a 1566 bp open reading frame resulting in a 521 amino acid protein. The Hsp40-L protein contains a J domain, zinc-finger domains, a glycine-phenylalanine rich region, but lacks cysteine-glycine repeat regions classifying it as a type II member of the J-protein family (Figure 2). It is most closely related to DNAJC21, the ortholog of jjj1 in yeast, which plays a role in ribosome biogenesis. Interestingly, this gene is polymorphic, with most of the polymorphisms found in the last 100 residues of the C-terminus. This corresponds to a region of the protein referred to as the client binding domain, which is thought to provide specificity for target proteins (reviewed in Kampinga and Craig, 2010). Although not nearly as diverse as cfuhcsec, there is statistical variation within populations as high as 80% (Nydam et al., 2013b). Polymorphisms of the hsp40-l also correlate with fusion/rejection outcomes (Shanley et al., in preparation). Through whole mount in situ hybridization using a probe against a unique portion of the gene, expression of the hsp40-l gene was detected in the epithelial cell layer of the ampullae (Nydam et al., 2013b). However, aside from being encoded within the fuhc locus and expressed at the site of allorecognition and an unusual distribution of polymorphism, no other data yet support its potential role in allorecognition.
2.4 BHF
Recently, Voskoboynik et al. (2013b) identified a new polymorphic gene involved in Botryllus allorecognition, Botryllus histocompatibility factor (bhf), which is located ca. 62 kb upstream of the secreted form of cfuhc and 95 kb downstream of the uncle fester locus (Figure 1). This was done using in silico analyses of polymorphisms detected following mRNA-seq of individuals of defined fusibility types that were independent of protein structure. They identified a 252 amino acid protein with no predicted conserved domains, that is partially unstructured and highly charged (Figure 2). Based on polymorphisms in the first 100 amino acids, bhf is able to predict the outcome of histocompatibility pairings. bhf is expressed in the ampullae and epithelium of vessels, reinforcing its potential role in allorecognition, and can also be amplified by RT-PCR from the blood, whole animal, endostyle and bud. Interestingly, qPCR showed an increase of bhf expression during the allorecognition response and higher mRNA expression compared to cfuhc. Morpholino-mediated knockdown and semi-quantitative PCR showed a maximum of ~70% knockdown by 48 hours post-injection and resulted in a ‘no reaction’ phenotype between fusing and rejecting pairs. The role of BHF in allorecognition remains unclear considering that this protein is predicted to localize to the cytoplasm, yet is also able to predict fusibility outcomes that are conventionally controlled by cell surface or extracellular proteins. Despite this, there are two hypotheses to explain the no-reaction phenotype. First, BHF is a ligand for both the rejection and fusion pathways, and is somehow transported to the outside of the cells it is expressed on. Second, BHF is involved in assembling or anchoring receptor complexes, which would be consistent with its unordered structure (reviewed in Uversky, 2011) and putative localization. However, it is difficult to interpret these experiments for several reasons. First, in previous knockdown experiments with fester and uncle fester, the ampullae were surgically removed and regenerated during siRNA treatment, such that siRNA was present when new protein was being synthesized in the regenerating ampullae, i.e., there were no issues of protein turnover. In addition, a lack of the corresponding protein was confirmed using immunofluorescence (McKitrick et al., 2011; Nyholm et al., 2006). Ampullae regeneration was not used in the BHF experiments, and it is unclear how a 2 day morpholino treatment could result in protein depletion, unless normal turnover of BHF protein is very high. In addition, in both experiments (fusing and rejecting pairs) only one of the two individuals in each pairing was under morpholino treatments. If BHF is a ligand, these results would suggest it is involved in a homotypic interaction for both fusion and rejection. Given that a no-reaction phenotype was seen in both compatible and incompatible pairings, these results are not consistent with either a missing-self model, or uncle fester knockdown results. In the latter, a rejection still occurred when a knockdown was paired with a control individual, and further that mAb stimulation of uncle fester could induce a rejection response in vivo (McKitrick et al., 2011).
In addition, the in silico analysis used to identify bhf and compare polymorphisms to alloreocognition outcomes should be viewed with some skepticism. Given that the cfuhcsec and bhf loci are located 67 Kb apart, it is unclear how they could have different predictive abilities, particularly since only 29 pairs of animals were analyzed. More importantly, the same analysis suggested that cfuhcsec and cfuhctm each had different predictive abilities, despite that fact that they are encoded only 227bp from each other. Finally, hsp40-l, which is encoded 8Kb from form cfuhctm, has significantly less predictive ability than those, and even unlinked genes. Given that the fuhc was recombination mapped on a population of several hundred individuals from multiple crosses, showed no hotspots of recombination, and delineated a region that is already over 1 Mb, it’s highly unlikely that the in silico analysis is correct. Given the sample size studied, it would suggest an improbable amount of recombination in the locus.
Despite these caveats, the discovery of bhf is an exciting finding, as the original studies of the fuhc locus only analyzed predicted genes with characteristics that would suggest their involvement in a histocompatibility reaction (De Tomaso et al., 2003, 2005). In addition to the finding of an hsp40-l which is similarly polymorphic, this suggests that intracellular proteins in this locus may also be important players in allorecognition and is currently being analyzed with the other genes.
3. Conclusion
In summary, polymorphisms of cfuhcsec, cfuhctm, hsp40-l and bhf have been shown to predict outcome of fusibility, which is not surprising given their tight linkage (ca. 110 Kb; Figure 1). Meanwhile, fester has been demonstrated to have roles in the fusion and rejection reactions, and it is possible that specificity is generated through numerous splice isoforms. Given that each individual from both lab-reared and wild-type displays different splice variant repertoires, this suggests how Fester may be involved in recognition of the putative cFuhc ligand (discussed below). uncle fester is solely involved in the rejection pathway, and this supports the hypothesis that the two reactions are not mutually exclusive, and there is an activating signal to induce a rejection. Only when a self marker is detected, an inhibitory signal will override the rejection response. Lastly, Hsp40-L is a chaperone protein with an unusual polymorphic C-terminal end that could be involved in allorecognition, most likely aiding in the folding or assembly of the previously mentioned players.
3.1 ‘Missing Self’ Model
B. schlosseri’s allorecognition system is reminiscent of the ‘missing self’ mechanism in NK cells, where the self marker is either one or multiple genes within the fuhc locus. Within a 110 kb region in this locus there are four genes (cfuhcsec, cfuhctm, bhf, hsp40-l) that predict allorecognition outcome along with two additional genes, fester and uncle fester, that can affect outcome based on functional data (McKitrick et al., 2011; Nyholm et al., 2006). Of course, without knowing the function of these four genes, any model proposed is speculative. However, to date the most polymorphic member of this group, and that with the most evidence for positive selection is cfuhcsec, and this would be the expected characteristics of the allodeterminant for a fester family receptor expressed along the epithelial cells of the ampullae.
3.1.1 As Colonies Touch
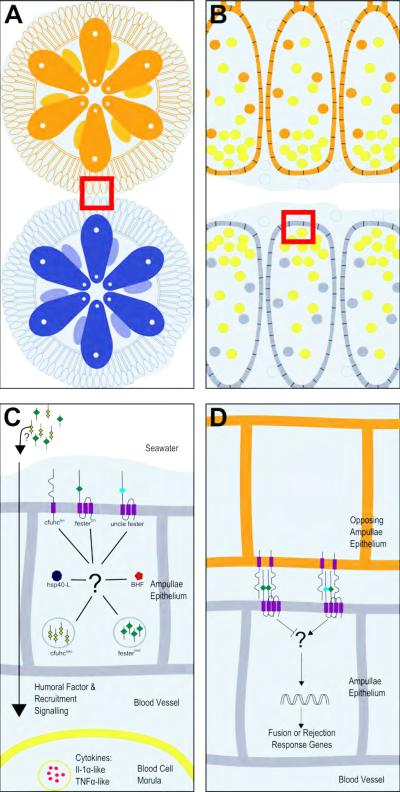
As described in the previous sections (1.2.1-1.2.3.2), when two individuals are near each other and their tunics come into contact (Figure 3A), morula cells, the effector cells of the rejection response, are observed to migrate and crowd into the ampullar tips (Figure 3B) regardless of histocompatibility outcome (Cima et al., 2004, 2006; Rinkevich et al., 1998). Cima et al. (2006) hypothesized that a good candidate for this chemoattractant is cFuhcsec. They speculated that in compatible pairs homing of morula cells to the ampullae tips is due to the unshared allele since ampullar populations of morula cells were higher in rejecting colonies compared to fusing. However, in our experience the morula cell infiltration is a variable phenotype, with some colonies reacting strongly, and others not at all, and does not appear to be correlated with compatibility. We also regularly observe morula cell migration in compatible pairings of fuhc homozygotes (unpublished). While cFuhcsec is an obvious candidate for early steps in allorecognition, it is not known where the protein is localized after being secreted. It may be interacting as a complex on the cell surface or diffusing into and through the tunic into the water, or in contrast be secreted in a polarized manner into the vasculature.
Figure 3.
Cartoon schematic of the initiation of allorecognition. (A) Two individuals (orange and blue) in close proximity of one another. Red box indicates potential interacting ampullae. (B) Magnified region of the red box in (A). Morula cells (yellow dots) are shown accumulating at the ampullar tips. Orange or blue dots represent pigment cells. (C) Magnified region of the red box in (B) with the candidate proteins. (D) A one-way directional portrayal of two interacting ampullae with the speculated candidates.
Alternatively, cFuhcsec could be the humoral factor that blocks the rejection response in morula cells but plays no role in their migration prior to POR formation (Figure 3C). This would be consistent with the observation that when morula cells are incubated with incompatible blood plasma, they will degranulate as they would during a rejection response (Ballarin et al., 1995, 1998; Cima et al., 2004, 2006). In contrast, when these cells were incubated with compatible blood plasma, no significant degranulation was observed.
Nydam et al. (2013a) demonstrated that multiple codons in the cfuhcsec are under positive selection (directional) compared to cfuhctm, which was less polymorphic. This suggests a role for individual specificity rather than a species-wide signal. In addition, if recognition of cfuhcsec is on the external membrane of the ampullae, then it could bind to its respective receptor both in cis and trans, making discrimination of the source of the secreted protein challenging. Similar arguments can be made for secreted Fester as a humoral factor since there are 16 possible splice isoforms along with polymorphic residues which are also under positive selection (Nydam and De Tomaso, 2012), and in fact this argument could be made for any diffusible factor. Since colonies are not in a constant state of inflammation, it is unclear how this diffusible signal is given, but most likely it is an increase in concentration near the juxtaposed ampullae that must cross a threshold to initiate the inflammatory response.
In a study of xenoreactions between 9 botryllid species, most species ignored each other. However, xeno-rejection was observed in several pairings without ampullae interactions while fusion never occurred (Hirose et al., 2002; Rinkevich et al., 1998; Saito, 2003). This indicates a somewhat conserved factor is activating the rejection response. Perhaps bhf could be this factor given its lower polymorphism and homologs in other tunicates (Voskoboynik et al., 2013b). While BHF is predicted to stay in the cytosol, it may interact with another protein to traffic it to the extracellular region through its disordered region (reviewed in Uversky, 2011). Potentially, the humoral factor activating morula cells in different botryllid species is the same, and in some cases the two can mimic each other, stimulating xenorejection. It would be of interest to examine if the other botryllids possess any similar genes, in addition to bhf, that are in the B. schlosseri fuhc locus. Along those lines, we have found homologs of all allorecognition proteins except fester and uncle fester in another botryllid found on the California coast, Botryllus tuberatus (Langenbacher et al., unpublished). Finally, it is also possible that there are conserved epitopes on the cfuhcsec which provide the activating signal.
In summary, it can be easily demonstrated that a diffusible signal alerts an individual to the presence of a nearby individual and activates an inflammatory response. This signal is usually species-specific, but not always. However, it is unclear how the source of a diffusible signal can be determined by any individual.
3.1.2 A Touching Moment
After fusion of the tunic and once opposing ampullae are in contact, the epithelial lining of the ampullae will be induced and become leaky, an activation step that will lead to a POR formation (rejection response). The receptor for this activation is probably composed of a Fester/Uncle Fester complex based on functional data (McKitrick et al., 2011; Nyholm et al., 2006). Whether or not this activating complex is purely a heterodimer or a mixture of homodimers of different splice isoforms of Fester or a homodimer of Uncle Fester remains to be determined. However, the activating complex should be able to bind any allele of the activating ligand (discussed above). The activation signaling that results in POR formation can be overturned when an inhibitory signal is sent after binding of a self ligand to the receptor. The receptor could be composed of different splice isoforms of fester that each recognizes self cFuhc ligands, with the avidity correlating to the strength of the inhibitory signal. The idea that fusion or rejection is ultimately due to interplay between two independent pathways controlling each outcome has been shown both directly and genetically. In two studies, fusion or rejection can be manipulated by stimulating the opposite pathway using mAbs to Fester and Uncle Fester (discussed above). In addition, the severity of the rejection response maps to the fuhc locus itself (Scofield and Nagashima, 1983), and a previous study correlated weak rejection to closely related, but non-matching cfuhcsec alleles (Nydam et al., 2013b). A weaker rejection may be explained by partial binding which would weakly stimulate an inhibitory pathway and lower the strength of the activating signal.
So how does an innate recognition system achieve the ability to identify a self-ligand from hundreds of competing specificities? On the surface of the epithelium, we hypothesize that there would be a population of Fester splice isoforms, each able to recognize some epitope on the self-ligand. These would be organized into isoform complexes that would bind such that an avidity of binding to self would be higher than to non-self. The formation of these complexes would involve Hsp40-L to help mediate folding, formation, and specificity in an education process, akin to stochastic expression of inhibitory receptors during mammalian NK-cell education. BHF could be functioning as an adaptor or a scaffolding protein in these complexes through its disordered region (reviewed in Uversky, 2011) since it is localized to the cytoplasm or perhaps in a complex with Hsp40-L to assist with its function. In summary, recognition of self ligands would be due to an overall avidity from binding of each fester splice variant. In turn, the splice variants would be assembled and anchored by Hsp40-L and BHF, in an education process that occurs during embryogenesis or metamorphosis, as newly hatched oozooids are able to discriminate between self and non-self (Scofield et al., 1982a).
From a purely topological viewpoint, the most likely ligand for these two receptors (activating and inhibitory) is cFuhctm given its expression profile and polymorphisms predicting outcome (Figure 3D). However, the same reasoning can be made for cFuhcsec - if it is interacting with another cell surface marker. BHF cannot be excluded either if it able to interact with an extracellular domain of a surface marker through its disordered region; thus providing a new mechanism of allorecognition as suggested by Voskoboynik et al. (2013b). Hsp40-L may also have a role in the processing of the ligand to the cell surface.
3.2 Challenges Ahead & Future Experiments
Over the past decade, steps to elucidate the allorecognition system and mechanism in B. schlosseri have been taken from identifying a putative ligand (De Tomaso et al, 2005; Nydam et al., 2013c), putative receptors (Nyholm et al., 2006; McKitrick et al., 2011) to other players (Nydam et al., 2013b; Voskoboynik et al., 2013b). However, with a developing model organism lacking some tools, for example transgenic lines, tackling questions such as the generation of specificity will be a challenge. At this point in time, the role and precise contribution of each gene to allorecognition are unknown, and extensive biochemical and molecular studies are in progress. This is difficult due to the fact that there are no tunicate cell lines which provide a robust in vitro model. Granted, there have been some genes from B. schlosseri expressed in a bacterial system: rhamnose-binding lectins (RBLs) (Gasparini, et al., 2008), CD94 (Khalturin et al., 2003), a C-type lectin (BSCLT) (Pancer et al., 1997). Meanwhile, fewer expressed genes have been biochemically investigated by extracting proteins directly from B. schlosseri: agglutinin (Ballarin et al., 1999), a heterodimer receptor (Danska et al., 1990), and phenoloxidase (Frizzo et al., 1999; Ballarin, et al., 2012).
Future experiments should focus on identifying the ligand involved in allorecognition. With potential candidates identified (cFuhcsec, cFuhctm, BHF), experiments involving stimulation assays using recombinant forms of each protein can elucidate the ligand. Additionally, knockdowns of cfuhcsec and cfuhctm need to be conducted to determine their functional involvement in allorecognition. However, due to the high amounts of polymorphisms across each gene, there have been many difficulties acquiring a repeatable knockdown, and efforts to create antibodies to these proteins have not yet been successful. Reciprocally, since putative receptors have been identified, pull-down assays can be conducted to identified ligands and other binding partners. This would address if Fester and Uncle Fester can form a heterodimer or not. Utilizing in vitro expression systems, individual allorecognition genes are being expressed to confirm localization and perhaps co-localization and binding interactions with other allorecognition genes in a systematic method that will also help address these questions. Lastly to help determine function, aside from in vivo tests such as the antibody interference experiments (McKitrick et al., 2011; Nyholm et al., 2006), a tissue culture based reporter system described in Mesci and Carlyle (2007) is being established to determine if binding between the putative ligands and receptors can induce signaling. While determining the biochemical and molecular roles of the allorecognition genes is not trivial, the results from these studies will prove to be invaluable to further defining and expanding our current knowledge of B. schlosseri allorecognition.
Finally, there has been no linkage with any allorecognition candidate proteins and intracellular signaling, as none of the genes described contain any conserved motifs. However, this has not prevented us from speculating that it is the signaling pathways themselves that are conserved, and at the core of specificity in allorecognition throughout the metazoan (reviewed in De Tomaso, 2009). This hypothesis is based on two observations. First, almost all mammalian immune function is due to integration of activating and inhibitory signals (kinases and phosphatases, respectively) from multiple, unrelated receptor systems that are rapidly evolving. The best example of this is seen in mammalian NK cells, in which activating and inhibitory receptors are in mice and primates use two completely different ectodomains (Ig in the KIR family in primates, C-type lectin in the Ly49 family in mice), but are still linked to the same intracellular signaling pathways, via intracellular ITIM and ITAM domains. Second, most of the signal transduction proteins in the ITIM and ITAM pathways are highly conserved, and found in the earliest metazoans. Thus it may be that it is intracellular events: e.g., signal integration and education processes that set the thresholds for appropriate responses to binding events at the cell surface that are the conserved portion of allorecognition. This could explain the rapid evolution of cell surface components observed in the evolution of allorecognition and immunity. Localization and functional studies of these proteins are also a priority.
Highlights.
We discuss the phenomenon of allorecognition in context of the evolution of immunity in the metazoa.
Recent findings on allorecognition in the colonial ascidian, Botryllus schlosseri are reviewed.
Potential models for allo-specificity in B. schlosseri are presented.
Acknowledgements
We thank Adam Langenbacher for helpful comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ballarin L, Cima F, Sabbadin A. Morula cells and histocompatibility in the colonial ascidian Botryllus schlosseri. Zool. Sci. 1995;12:757–764. [Google Scholar]
- Ballarin L, Cima F, Sabbadin A. Phenoloxidase and cytotoxicity in the compound ascidian Botryllus schlosseri. Dev. Comp. Immunol. 1998;22:479–492. doi: 10.1016/s0145-305x(98)00035-4. [DOI] [PubMed] [Google Scholar]
- Ballarin L, Tonello C, Guidolin L, Sabbadin A. Purification and characterization of a humoral opsonin, with specificity for D-galactose, in the colonial ascidian Botryllus schlosseri. Comp. Biochem. Physiol. 1999;123B:115–123. doi: 10.1016/s0305-0491(99)00050-4. [DOI] [PubMed] [Google Scholar]
- Ballarin L, Cima F, Floreani M, Sabbadin A. Oxidative stress induces cytotoxicity during rejection reaction in the compound ascidian Botryllus schlosseri. Comp Biochem Physiol C Toxicol Pharmacol. 2002;133:411–418. doi: 10.1016/s1532-0456(02)00123-0. [DOI] [PubMed] [Google Scholar]
- Ballarin L, Franchi N, Schiavon F, Tosatto SCE, Mičetić I, Kawamura K. Looking for putative phenoloxidases of compound ascidians: Haemocyanin-like proteins in Polyandrocarpa misakiensis and Botryllus schlosseri. Dev. Comp. Immunol. 2012;38:232–242. doi: 10.1016/j.dci.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Bancroft FW. Variation and fusion of colonies in compound ascidians. Proc. Calif. Acad. Sci. 1903;3:137–186. 3rd ser. [Google Scholar]
- Boyd HC, Weissman IL, Saito Y. Morphologic and genetic verification that Monterey Botryllus and Woods Hole Botryllus are the same species. Biol. Bull. 1990;178:239–250. doi: 10.2307/1541825. [DOI] [PubMed] [Google Scholar]
- Brown FD, Tiozzo S, Roux MM, Ishizuka K, Swalla BJ, De Tomaso AW. Early lineage specification of long-lived germline precursors in the colonial ascidian Botryllus schlosseri. Development. 2009;136:3485–3494. doi: 10.1242/dev.037754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnet FM. Self-recognition” in colonial marine forms and flowering plants in relation to the evolution of immunity. Nature. 1971;232:230–235. doi: 10.1038/232230a0. [DOI] [PubMed] [Google Scholar]
- Buss LW. Somatic cell parasitism and the evolution of somatic tissue compatibility. Proc. Natl. Acad. Sci. 1982;79:5337–5341. doi: 10.1073/pnas.79.17.5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cima F, Sabbadin A, Ballarin L. Cellular aspects of allorecognition in the compound ascidian Botryllus schlosseri. Dev. Comp. Immunol. 2004;28:881–889. doi: 10.1016/j.dci.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Cima F, Sabbadin A, Zaniolo G, Ballarin L. Colony specificity and chemotaxis in the compound ascidian Botryllus schlosseri. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2006;145:376–382. doi: 10.1016/j.cbpa.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Danska JS, McIntrye BW, McDevitt HO, Weissman IL. Structural similarity between a primitive chordate membrane heterodimer and lymphocyte antigen receptors. Int. Immunol. 1990;2(9):795–802. doi: 10.1093/intimm/2.9.795. [DOI] [PubMed] [Google Scholar]
- De Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. ScanProsite: detection of PROSTIE signature matches and ProFule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–5. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are not the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- De Tomaso AW, Saito Y, Ishizuka KJ, Palmeri KJ, Weissman IL. Mapping the genome of a model protochordate. I. A low resolution genetic map encompassing the fusion/histocompatibility (Fu/HC) locus of Botryllus schlosseri. Genetics. 1998;149:277–287. doi: 10.1093/genetics/149.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Tomaso AW, Weissman IL. Initial characterization of a protochordate histocompatibility locus. Immunogenetics. 2003a;55:480–490. doi: 10.1007/s00251-003-0612-7. [DOI] [PubMed] [Google Scholar]
- De Tomaso AW, Weissman IL. Construction and characterization of large-insert genomic libraries (BAC and fosmid) from the ascidian Botryllus schlosseri and initial physical mapping of a histocompatibility locus. Mar. Biotechnol. 2003b;5:103–115. doi: 10.1007/s10126-002-0071-4. [DOI] [PubMed] [Google Scholar]
- De Tomaso AW, Nyholm SV, Palmeri KJ, Ishizuka KJ, Ludington WB, Mitchel K, Weissman IL. Isolation and characterization of a protochordate histocompatibility locus. Nature. 2005;438:454–459. doi: 10.1038/nature04150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Tomaso AW. Sea squirts and immune tolerance. Dis. Models Mech. 2009;2:440–445. doi: 10.1242/dmm.001156. [DOI] [PubMed] [Google Scholar]
- Fagan MB, Weissman IL. Sequence and characterization of two HSP70 genes in the colonial protochordate Botryllus schlosseri. Immunogenetics. 1996;44:134–142. doi: 10.1007/BF02660062. [DOI] [PubMed] [Google Scholar]
- Frizzo A, Guidolin L, Ballarin L, Sabbadin A. Purification and partial characterisation of phenoloxidase from the colonial ascidian Botryllus schlosseri. Mar. Biol. 1999;135:483–488. [Google Scholar]
- Gasparini F, Franchi N, Spolaore B, Ballarin L. Novel rhamnose-binding lectins from the colonial ascidian Botryllus schlosseri. Dev. Comp. Immunol. 2008;32:1177–1191. doi: 10.1016/j.dci.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Grosberg RK, Quinn JF. The genetic control and consequences of kin recognition by the larvae of a colonial marine invertebrate. Nature. 1986;322:456–459. [Google Scholar]
- Hastings KEM. SL trans-splicing: easy come or easy go? Trends Genet. 2005;21(4):240–247. doi: 10.1016/j.tig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Hirose E, Shirae M, Saito Y. Colony specificity in the xenogeneic combinations among four Botrylloides species (Urochordata, Ascidiacea) Zool. Sci. 2002;19(7):747–753. doi: 10.2108/zsj.19.747. [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalturin K, Becker M, Rinkevich B, Bosch TCG. Urochordates and the origin of natural killer cells: Identification of a CD94/NKR-P1-related receptor in blood cells of Botryllus. Proc. Natl. Acad. Sci. 2003;100(2):622–627. doi: 10.1073/pnas.0234104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird DJ, De Tomaso AW, Weissman IL. Stem cells are units of natural selection in a colonial ascidian. Cell. 2005;123:1351–1360. doi: 10.1016/j.cell.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:302–305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi L, Douek J, Osman M, Bosch TCG, Rinkevich B. Cloning and characterization of BS-cadherin, a novel cadherin from the colonial urochordate Botryllus schlosseri. Gene. 1997;200:117–123. doi: 10.1016/s0378-1119(97)00391-0. [DOI] [PubMed] [Google Scholar]
- McKitrick TR, Muscat CC, Pierce JD, Bhattacharya D, De Tomaso AW. Allorecognition in a basal chordate consists of independent activating and inhibitory pathways. Immunity. 2011;34:616–626. doi: 10.1016/j.immuni.2011.01.019. [DOI] [PubMed] [Google Scholar]
- Mesci A, Carlyle JR. A rapid and efficient method for the generation and screening of monoclonal antibodies specific for cell surface antigens. J. Immunol. Methods. 2007;323(1):78–87. doi: 10.1016/j.jim.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Menin A, del Favero M, Cima F, Ballarin L. Release of phagocytosis-stimulating factor(s) by morula cells in a colonial ascidian. Marine Biol. 2005;148:225–230. [Google Scholar]
- Milanesi C, Burighel P, Zaniolo G, Sabbadin A. The structure and the fate of the test cuticle during the fusion-nonfusion reaction in the colonies of Botryllus schlosseri (Tunicata) Boll. Zool. 1978;45:83–86. [Google Scholar]
- Nydam ML, De Tomaso AW. The fester locus in Botryllus schlosseri experiences selection. BMC Evol. Biol. 2012;12:249. doi: 10.1186/1471-2148-12-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nydam ML, Taylor AA, De Tomaso AW. Evidence for selection on a chordate histocompatibility locus. Evolution. 2013a;67(2):487–500. doi: 10.1111/j.1558-5646.2012.01787.x. [DOI] [PubMed] [Google Scholar]
- Nydam ML, Hoang TA, Shanley KM, De Tomaso AW. Molecular evolution of a polymorphic HSP40-like protein encoded in the histocompatibility locus of an invertebrate chordate. Dev. Comp. Immunol. 2013b;41:128–136. doi: 10.1016/j.dci.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Nydam ML, Netuschil N, Sanders E, Langenbacher A, Lewis DD, Taketa DA, Marimuthu A, Gracey AY, De Tomaso AW. The candidate histocompatibility locus of a basal chordate encodes two highly polymorphic proteins. PLoS ONE. 2013c;8(6):e65980. doi: 10.1371/journal.pone.0065980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Passegue E, Ludington WB, Voskoboynik A, Mitchel K, Weissman IL, De Tomaso AW. fester, a candidate allorecognition receptor from a primitive chordate. Immunity. 2006;25:163–173. doi: 10.1016/j.immuni.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Oka H, Watanabe H. Colony-specificity in compound ascidians as tested by fusion experiments. Proc. Jpn. Acad. Sci. 1957;33:657–664. [Google Scholar]
- Oren M, Douek J, Fishelson Z, Rinkevich B. Identification of immune-relevant genes in histoincompatible rejecting colonies of tunicate Botryllus schlosseri. Dev. Comp. Immunol. 2007;31:889–902. doi: 10.1016/j.dci.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Oren M, Escande ML, Paz G, Fishelson Z, Rinkevich B. Urochordate histoincompatible interactions activate vertebrate-like coagulation system components. PLoS ONE. 2008;3(9):e3123. doi: 10.1371/journal.pone.0003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancer Z, Gershon H, Rinkevich B. cDNA cloning of a putative protochordate FK506-binding protein. Biochem. Biophys. Res. 1993;197(2):973–977. doi: 10.1006/bbrc.1993.2574. [DOI] [PubMed] [Google Scholar]
- Pancer Z, Gershon H, Rinkevich B. Cloning of a urochordate cDNA feature mammalian short consensus repeats (SCR) of complement-control protein superfamily. Comp. Biochem. Physiol. 1995a;111B(4):625–632. doi: 10.1016/0305-0491(95)00025-4. [DOI] [PubMed] [Google Scholar]
- Pancer Z, Gershon H, Rinkevich B. Coexistence and possible parasitism of somatic and germ cell lines in chimeras of the colonial urochordate Botryllus schlosseri. Biol. Bull. 1995b;189:106–112. doi: 10.2307/1542460. [DOI] [PubMed] [Google Scholar]
- Pancer Z, Scheffer U, Müller I, Müller WEG. Cloning of sponge (Geodia cydonium) and tunicate (Botryllus schlosseri) proteasome subunit epsilon (PRCE): Implications about the vertebrate MHC-encoded homologue LMP7 (PRCC) Biochem. Biophys. Res. Commun. 1996 doi: 10.1006/bbrc.1996.1674. [DOI] [PubMed] [Google Scholar]
- Pancer Z, Diehl-Seifert B, Rinkevich B, Müller WEG. A novel tunicate (Botryllus schlosseri) putative C-type lectin features an immunoglobulin domain. DNA and Cell Biology. 1997;16(6):801–806. doi: 10.1089/dna.1997.16.801. [DOI] [PubMed] [Google Scholar]
- Peterson TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Rinkevich B, Weissman IL. Botryllus schlosseri (Tunicata) whole colony irradiation: do senescent zooid resorption and immunological resorption involve similar recognition events? J. Exp. Zool. 1990;253:189–201. doi: 10.1002/jez.1402530209. [DOI] [PubMed] [Google Scholar]
- Rinkevich B, Weissman IL. Interpopulation allogenic reactions in the colonial protochordate Botryllus schlosseri. Int. Immunol. 1991;3(12):1265–1272. doi: 10.1093/intimm/3.12.1265. [DOI] [PubMed] [Google Scholar]
- Rinkevich B, Shapira M, Weissman IL, Saito Y. Allogenic responses between three remote populations of the cosmopolitan ascidian Botryllus schlosseri. Zool. Sci. 1992;9:989–994. [Google Scholar]
- Rinkevich B, Porat R, Goren M. Allorecognition elements on a urochordate histocompoatibility locus indicated unprecedented extensive polymorphism. Proc. R. Soc. Lond. 1995;259B:319–324. [Google Scholar]
- Rinkevich B, Tartakover S, Gershon H. Contribution of morula cells to allogenic responses in the colonial urochorodate Botryllus schlosseri. Marine BIol. 1998;131:227–236. [Google Scholar]
- Rinkevich B, Paz G, Douek J, Ben-Shlomo R. Allorecognition and microsatellite allele polymorphism of Botryllus schlosseri from the Adriatc sea. In: Sawada H, Yokosawa H, Lambert CC, editors. The Biology of Ascidians. Springer-Verlag; Tokyo: 2001. pp. 426–435. [Google Scholar]
- Rinkevich B, Douek J, Rabinowitz C, Paz G. The candidate Fu/HC gene in Botryllus schlosseri (Urochordata) and ascidians’ historecognition – An oxymoron? Dev. Comp. Immunol. 2012;36:718–727. doi: 10.1016/j.dci.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Rosner A, Rabinowitz C, Moiseeva E, Voskoboynik A, Rinkevich B. BS-Cadherin in colonial urochordate Botryllus schlosseri: One protein, many functions. Dev. Biol. 2007;304:687–700. doi: 10.1016/j.ydbio.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Sabbadin A. Le basi genetiche dell capacita di fusione fra colonie in Botryllus schlosseri (Ascidiacea) Atti Accad. Naz. Lincei Rend. 1962;32:1031–1035. [Google Scholar]
- Sabbadin A, Zaniolo G. Sexual differentiation and germ cell transfer in the colonial ascidian Botryllus schlosseri. J. Exp. Zool. 1979;207:289–304. [Google Scholar]
- Sabbadin A, Astorri C. Chimeras and histocompatibility in the colonial ascidian Botryllus schlosseri. Dev. Comp. Immunol. 1988;12:737–747. doi: 10.1016/0145-305x(88)90049-3. [DOI] [PubMed] [Google Scholar]
- Sabbadin A, Zaniolo G, Ballarin L. Genetic and cytological aspects of histocompatibility in ascidians. Boll. Zool. 1992;59:167–173. [Google Scholar]
- Saito Y, Watanabe H. Partial biochemical characterization of humoral factors involved in the nonfusion reaction of a botryllid ascidian, Botrylloides simodensis. Zool. Sci. 1984;1:229–235. [Google Scholar]
- Saito Y. Xenogeneic rejection among three botryllids (compound ascidians) Zool. Sci. 2003;20(5):481–589. doi: 10.2108/zsj.20.581. [DOI] [PubMed] [Google Scholar]
- Scofield VL, Schlumpberger JM, West LA, Weissman IL. Protochordate allorecognition is controlled by a MHC-like gene system. Nature. 1982a;295:499–502. doi: 10.1038/295499a0. [DOI] [PubMed] [Google Scholar]
- Scofield VL, Schlumpberger JM, Weissman IL. Colony specificity in the colonial tunicate Botryllus and the origins of vertebrate immunity. Amer. Zool. 1982b;22:783–794. [Google Scholar]
- Scofield VL, Nagashima LS. Morphology and genetics of rejection reactions between oozooids from the tunicate Botryllus schlosseri. Biol. Bull. 1983;165:733–744. doi: 10.2307/1541475. [DOI] [PubMed] [Google Scholar]
- Stoner DS, Weissman IL. Somatic and germ cell parasitism in a colonial ascidian: Possible role for a highly polymorphic allorecognition system. Proc. Natl. Acad. Sci. 1996;93:15254–15259. doi: 10.1073/pnas.93.26.15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner DS, Rinkevich B, Weissman IL. Heritable germ and somatic cell lineage competitions in chimeric colonial protochordates. Proc. Natl. Acad. Sci. 1999;96:9148–9153. doi: 10.1073/pnas.96.16.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneda Y, Watanabe H. Studies on colony specificity in the compound ascidian, Botryllus primigenus oka. I. Initiation of “nonfusion” reaction with special reference to blood cells infiltration. Dev. Comp. Immunol. 1982;6:43–52. doi: 10.1016/0145-305x(82)90006-4. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Intrinsically disordered proteins from A to Z. IJBCB. 2011;43:1090–1103. [Google Scholar]
- Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S. Innate or adaptive immunity? The example of Natural Killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskoboynik A, Neff NF, Sahoo D, Newman AM, Pushkarev D, Koh W, Passarelli B, Fan HC, Mantalas GL, Palmeri KJ, Ishizuka KJ, Gissi C, Griggio F, Ben-Shlomo R, Corey DM, Penland L, White RA, Weissman IL, Quake SR. The genome sequence of the colonial chordate, Botryllus schlosseri. eLife. 2013a;2:e00569. doi: 10.7554/eLife.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskoboynik A, Newman AM, Corey DM, Sahoo D, Pushkarev D, Neff NF, Passarelli B, Koh W, Ishizuka KJ, Palmeri KJ, Dimov IK, Keasar C, Fan HC, Mantalas GL, Sinha R, Penland L, Quake SR, Weissman IL. Identification of a colonial chordate histocompatibility gene. Science. 2013b;341:384–387. doi: 10.1126/science.1238036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman IL, Saito Y, Rinkevich B. Allorecognition histocompatibility in a protochordate species: Is the relationship to MHC somatic or structural? Immunol. Rev. 1990;113:227–241. doi: 10.1111/j.1600-065x.1990.tb00043.x. [DOI] [PubMed] [Google Scholar]