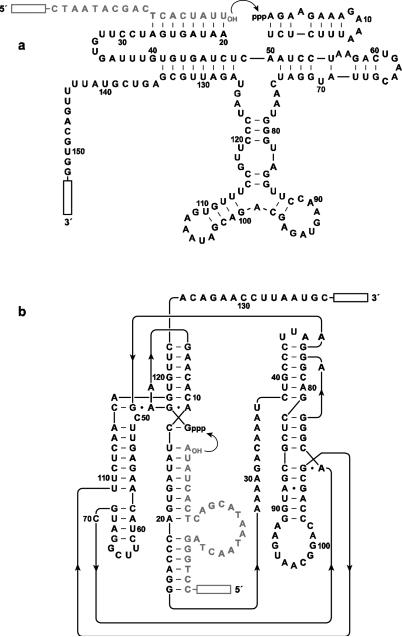
Fig 2.
Sequence and secondary structure of ribozymes used to initiate neutral drift experiments. a DSL ligase variant that wasselected for improved activity under low-Mg2+conditions.b Class I ligase variant that emerged from prior in vitro evolution experiments (Paegel and Joyce 2008). The substrate is shown in gray; curved arrow indicates the ligation junction; boxed regions indicate primer sites for reverse transcription and PCR amplification. Nucleotides that were subject to evolution are numbered.