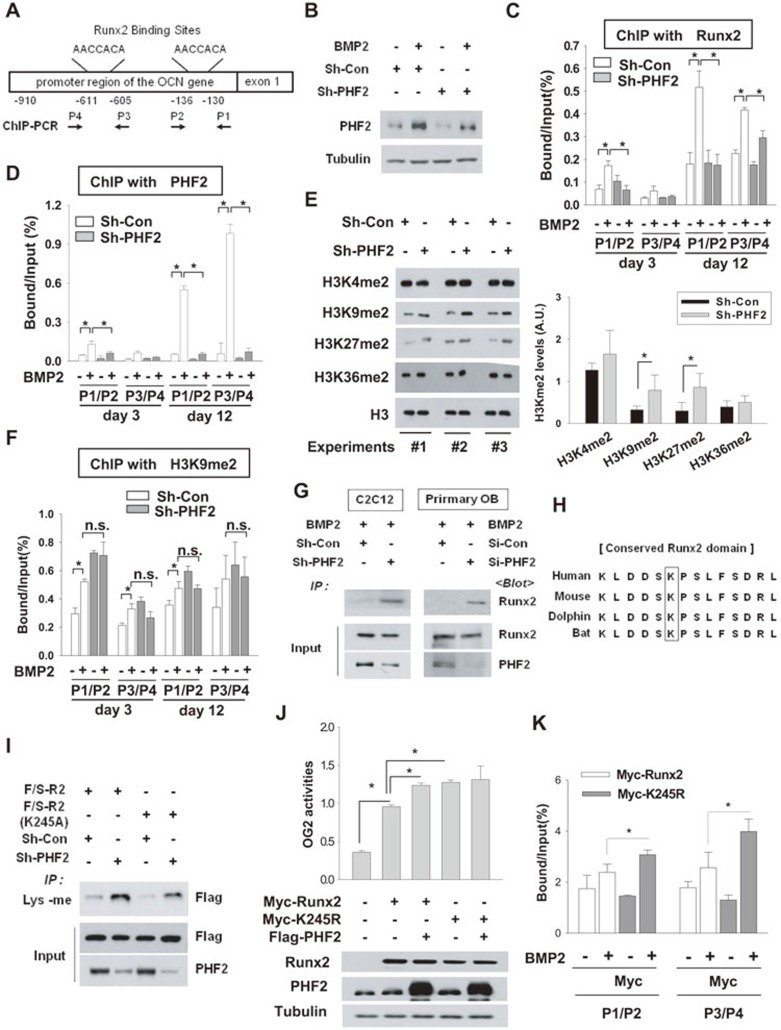
Figure 6.
PHF2 demethylated Runx2 and promoted the binding of Runx2 to DNA. (A) A schematic representation of OCN promoter and segments amplified by PCR. (B) C2C12 stable cell lines were incubated with BMP2 for 12 days, and PHF2 was immunoblotted. (C, D) ChIP for the binding of Runx2 or PHF2 to OCN promoter. C2C12 stable cell lines were incubated with BMP2 for 12 days. Chromatin was cross-linked and immunoprecipitated using anti-Runx2 (C) or anti-PHF2 (D). Precipitated DNAs were amplified and quantified by real-time PCR. The results (mean ± SD, n = 3 or more) are expressed as percentages of the input level. (E) The levels of dimethylated H3K4, H3K9, H3K27, H3K36, and H3 were analyzed by immunoblotting in C2C12 stable cell lines. (F) ChIP was performed six times with anti-H3K9me2 antibody in C2C12 stable cell lines. * and n.s. denote P < 0.05 and non-significant, respectively. (G) C2C12 stable cell lines or primary osteoblasts, which had been transfected with control or PHF2 siRNA, were treated with BMP2 for 24 h. Lysyl-methylated Runx2 was precipitated using anti-methylated lysine (Lys-me), and precipitated Runx2 was immunoblotted. (H) Comparisons of Runx2 sequences among human, mouse, dolphin, and bat. (I) C2C12 stable cell lines transfected with the F/S-R2 or F/S-R2 (K245A) plasmid were subjected to immunoprecipitation with anti-Lys-me, and the precipitated F/S-R2 peptides were immunoblotted. (J) C2C12 cells were co-transfected with Myc-Runx2 (or its K245R mutant), OG2 promoter-luciferase reporter, Flag-PHF2, and β-gal plasmids. The results indicate the mean ± SD (n = 4). (K) ChIP was performed to identify the binding of Myc-Runx2 and Myc-Runx2 (K245R) to OCN promoter. Transfected C2C12 cells were incubated with BMP2 for 48 h. ChIP was done with anti-Myc, and the precipitated DNA fragments were analyzed by real-time PCR. The results (mean ± SD, n = 3) are expressed as percentages of the input level. * denotes P < 0.05.