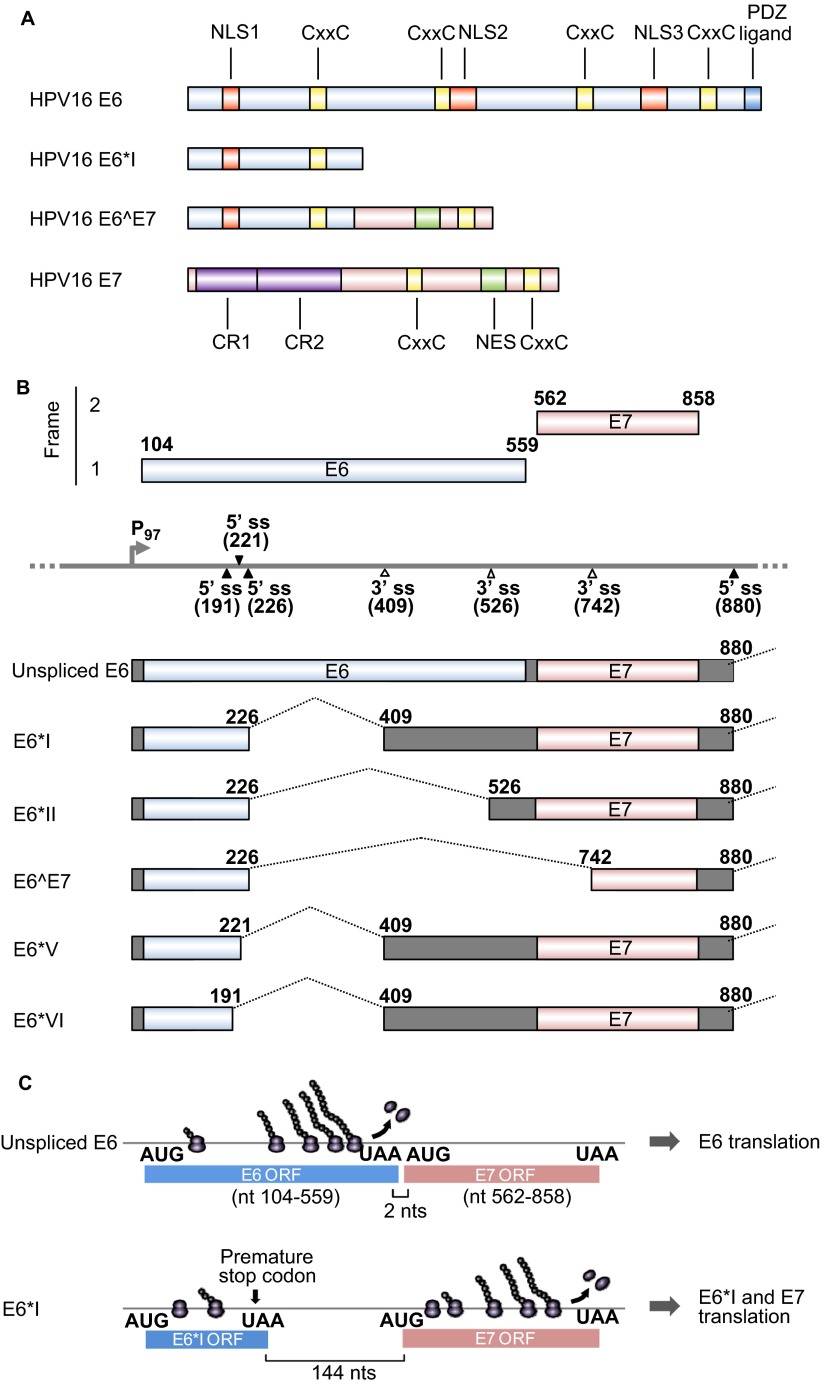
Figure 1.
HPV16 E6 and E7 and alternative RNA splicing. (A) Major functional domains and motifs of the HPV16 E6, E6*I, E6̂E7 and E7 proteins. HPV16 E6̂E7 has the N-terminal half of E6 and the C-terminal half of E7. (B) Alternative RNA splicing products of HPV16 E6E7 pre-mRNA. Alternative RNA splicing (dashed lines) takes place from three 5′ ss at nt 191, 221 and 226 to three alternative 3′ ss at nt 409, 526 and 742.27 The majority of RNA splicing occurs from the nt 226 5′ ss to the nt 409 3′ ss to produce E6*I, which is responsible for E7 translation.28 E6*III derived from the nt (226 5′ ss to the nt 3358 3′ ss and E6*IV derived from the nt 226 5′ ss to the nt 2709 3' ss29 are not included in this diagram. (C) Illustration of a ribosomal scanning model in HPV16 E6 and E7 translation, which is regulated by RNA splicing. Full-length E6 is translated from the unspliced E6 mRNA (upper diagram). E7 (nt 562–858) is translated from spliced E6*I mRNA in which a premature stop codon (UAA) is introduced by RNA splicing to form the E6*I ORF (lower diagram), which enlarges the space between the two ORFs. This enables a scanning ribosome to terminate translation of E6*I and reinitiate translation of E7. Nucleotide positions are numbered according to the HPV16 reference genome (PaVE, http://pave.niaid.nih.gov).