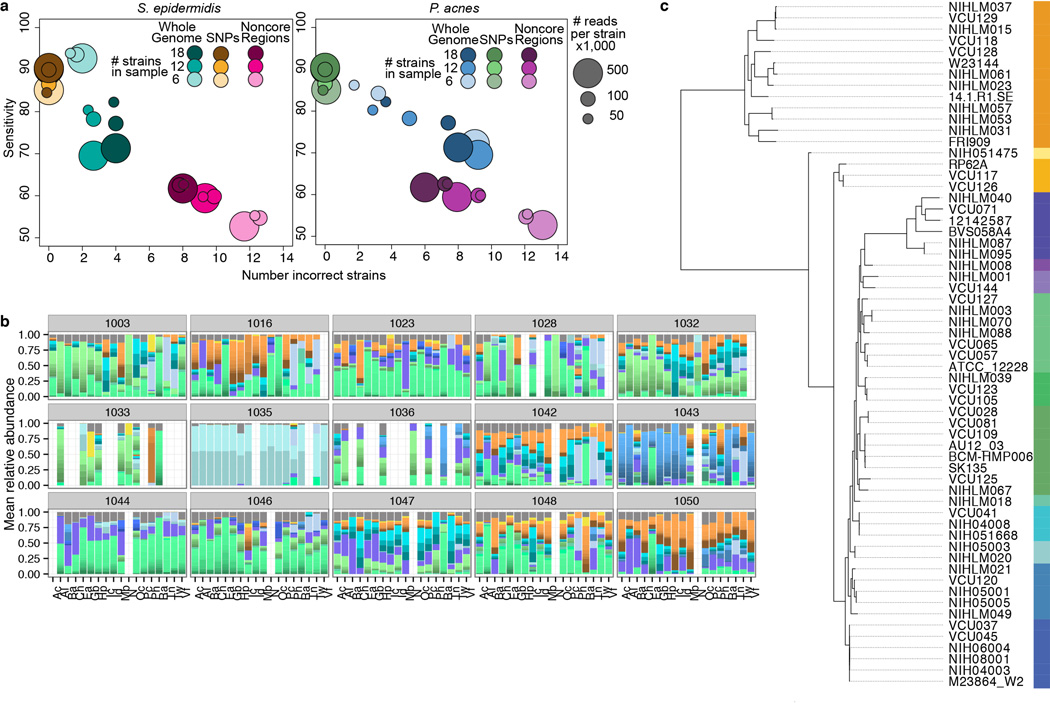
Extended Data Figure 5.
Strain-level classification based on reference genomes show sub-species heterogeneity for dominant skin taxa. a, Simulations to assess sensitivity of Pathoscope-based mapping to SNPs, non-core regions, or whole genomes. Synthetic communities were created with 6, 12, or 18 genomes per community. Sizes of circles reflect the number of reads sampled from each genome, e.g., 50,000, 1000,000, or 500,000 reads per genome. 15 random synthetic communities for each genome group were created and mapped to SNPs, non-core regions, or the full genome set. Sensitivity is calculated from the expected vs. the observed abundances. b, Full strain-level assignments for samples with relative abundances of closest related Propionibacterium acnes strains, by individual. c, Dendrograms of strain similarity. Trees were generated using core SNPs; genomes were aligned with nucmer to identify core regions, and then SNPs within these core regions were identified by calculating all pairwise differences between genomes. Bar of colors indicates delineations of subtypes where phylogenetically more similar genomes are in similar colors, e.g., we defined 12 subtypes for P. acnes.